"zinc finger, RAN-binding domain containing 3"
ZFIN 
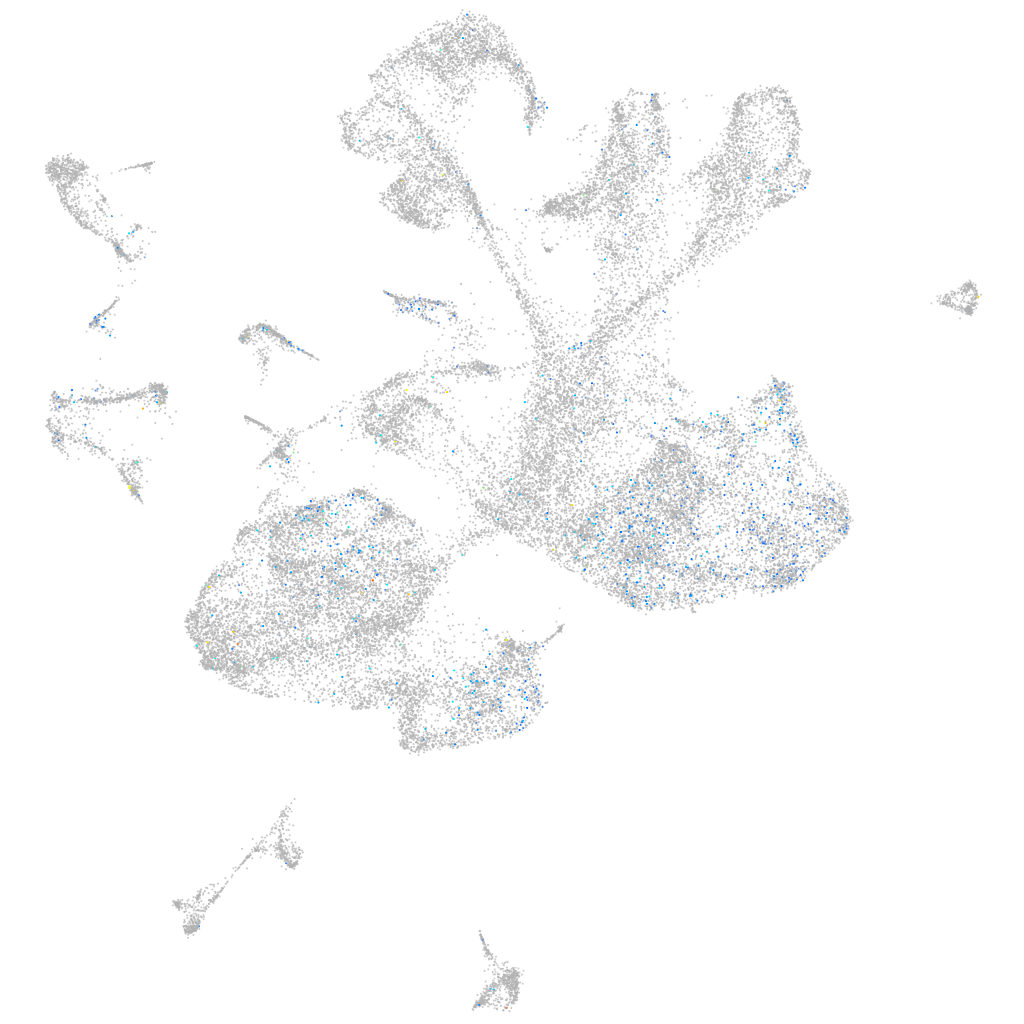
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| pcna | 0.124 | elavl3 | -0.061 |
| mibp | 0.122 | rtn1a | -0.060 |
| dut | 0.118 | myt1b | -0.058 |
| rrm1 | 0.114 | ppdpfb | -0.052 |
| zgc:110540 | 0.114 | stmn1b | -0.050 |
| rrm2 | 0.114 | gpm6ab | -0.049 |
| rpa2 | 0.112 | gpm6aa | -0.048 |
| esco2 | 0.109 | gng3 | -0.048 |
| lig1 | 0.109 | ckbb | -0.047 |
| chaf1a | 0.108 | tmsb | -0.047 |
| dek | 0.107 | rnasekb | -0.046 |
| banf1 | 0.107 | si:dkeyp-75h12.5 | -0.046 |
| zgc:110216 | 0.106 | dpysl3 | -0.044 |
| fbxo5 | 0.106 | marcksl1b | -0.043 |
| si:dkey-261m9.17 | 0.105 | sncb | -0.043 |
| rpa3 | 0.105 | elavl4 | -0.043 |
| cks1b | 0.104 | vamp2 | -0.043 |
| ccna2 | 0.103 | stx1b | -0.042 |
| CABZ01005379.1 | 0.103 | ube2d4 | -0.042 |
| nasp | 0.103 | atp6v0cb | -0.041 |
| fen1 | 0.102 | stxbp1a | -0.041 |
| prim2 | 0.100 | CR383676.1 | -0.041 |
| dhfr | 0.099 | calm1a | -0.040 |
| unga | 0.097 | snap25a | -0.040 |
| rnaseh2a | 0.097 | ywhag2 | -0.039 |
| suv39h1b | 0.096 | map1aa | -0.039 |
| slbp | 0.094 | zc4h2 | -0.039 |
| cdca5 | 0.094 | atp6v1e1b | -0.039 |
| rfc5 | 0.094 | cotl1 | -0.039 |
| stmn1a | 0.093 | scrt2 | -0.039 |
| LOC100330864 | 0.093 | syt2a | -0.039 |
| nutf2l | 0.093 | gng2 | -0.038 |
| smc2 | 0.093 | aplp1 | -0.038 |
| lbr | 0.093 | stmn2a | -0.038 |
| si:dkey-6i22.5 | 0.092 | atp6v1g1 | -0.038 |