zinc finger protein 536
ZFIN 
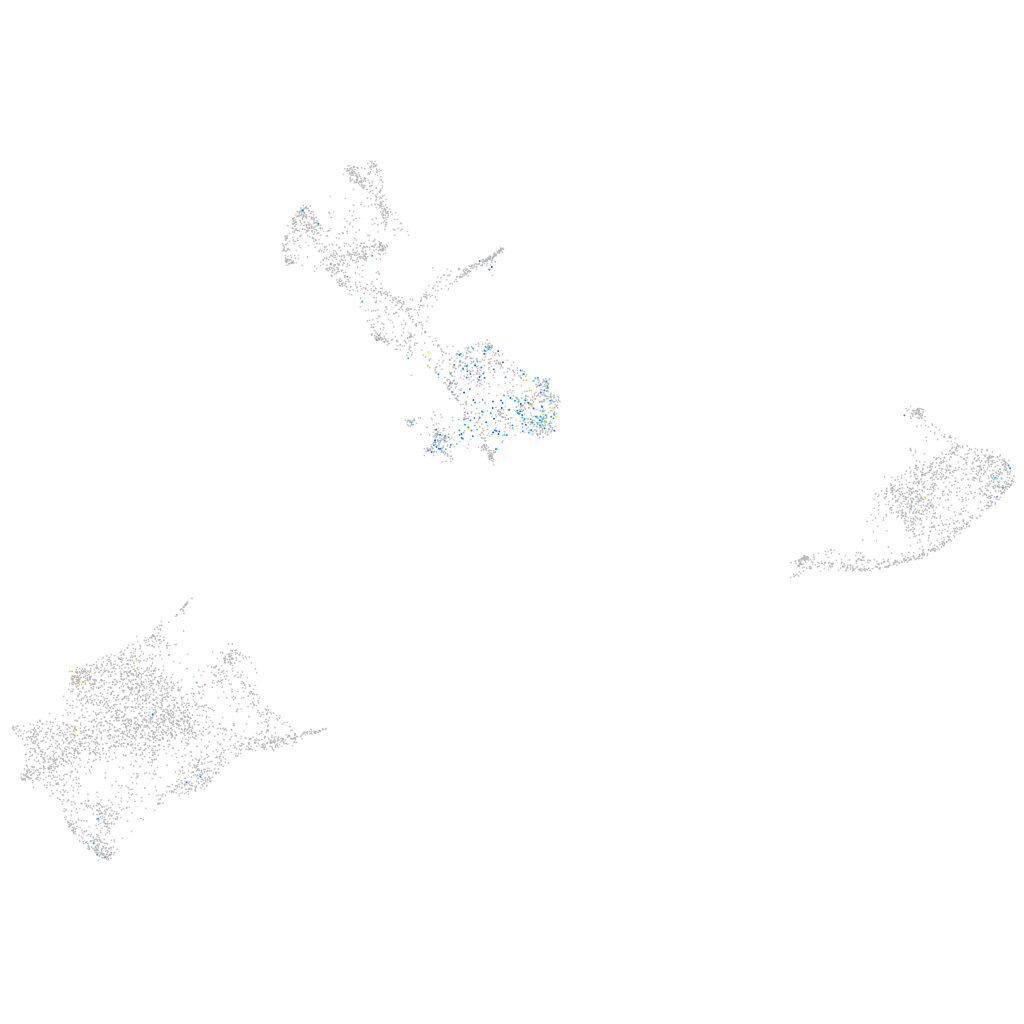
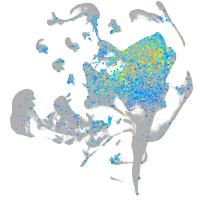
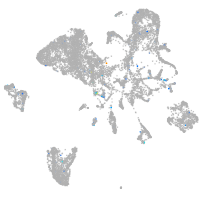
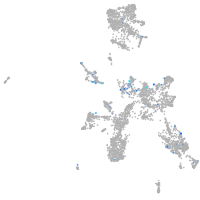
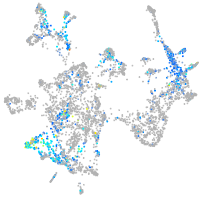
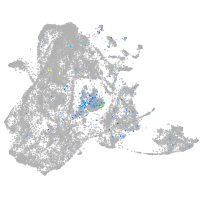
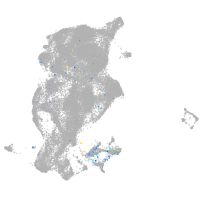
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| stmn1b | 0.267 | paics | -0.094 |
| nova2 | 0.255 | rab32a | -0.092 |
| elavl3 | 0.252 | akr1b1 | -0.092 |
| gpm6aa | 0.242 | gstp1 | -0.091 |
| tuba1c | 0.234 | impdh1b | -0.087 |
| tmeff1b | 0.227 | tspan36 | -0.085 |
| maptb | 0.226 | gpr143 | -0.083 |
| nat8l | 0.224 | pcbd1 | -0.083 |
| sncb | 0.224 | syngr1a | -0.083 |
| rtn1a | 0.221 | pts | -0.080 |
| hmgb1b | 0.221 | smim29 | -0.080 |
| gng3 | 0.220 | rnaseka | -0.077 |
| CU467822.1 | 0.217 | atic | -0.076 |
| zc4h2 | 0.217 | glulb | -0.076 |
| stmn2a | 0.217 | gpd1b | -0.075 |
| elavl4 | 0.216 | pttg1ipb | -0.075 |
| bcl11ba | 0.215 | rab38 | -0.074 |
| rnasekb | 0.214 | gch2 | -0.073 |
| gabrb4 | 0.212 | trpm1b | -0.073 |
| ptmaa | 0.211 | eno3 | -0.072 |
| fez1 | 0.209 | si:dkey-21a6.5 | -0.072 |
| tmsb | 0.209 | gmps | -0.072 |
| mllt11 | 0.208 | qdpra | -0.072 |
| map1aa | 0.207 | anxa4 | -0.069 |
| cacnb4b | 0.203 | rpl12 | -0.069 |
| cdk5r1b | 0.203 | rab34b | -0.069 |
| tubb5 | 0.203 | pfn1 | -0.069 |
| mpped2 | 0.203 | zgc:165573 | -0.068 |
| neurod6b | 0.202 | C12orf75 | -0.067 |
| cadm4 | 0.200 | actb2 | -0.066 |
| hmgb3a | 0.199 | rabl6b | -0.066 |
| olfm2a | 0.198 | gart | -0.066 |
| map2 | 0.196 | CABZ01032488.1 | -0.065 |
| nrxn1a | 0.196 | tmem243a | -0.065 |
| ppp1r14ba | 0.196 | CABZ01048402.2 | -0.065 |