zinc finger protein 516
ZFIN 
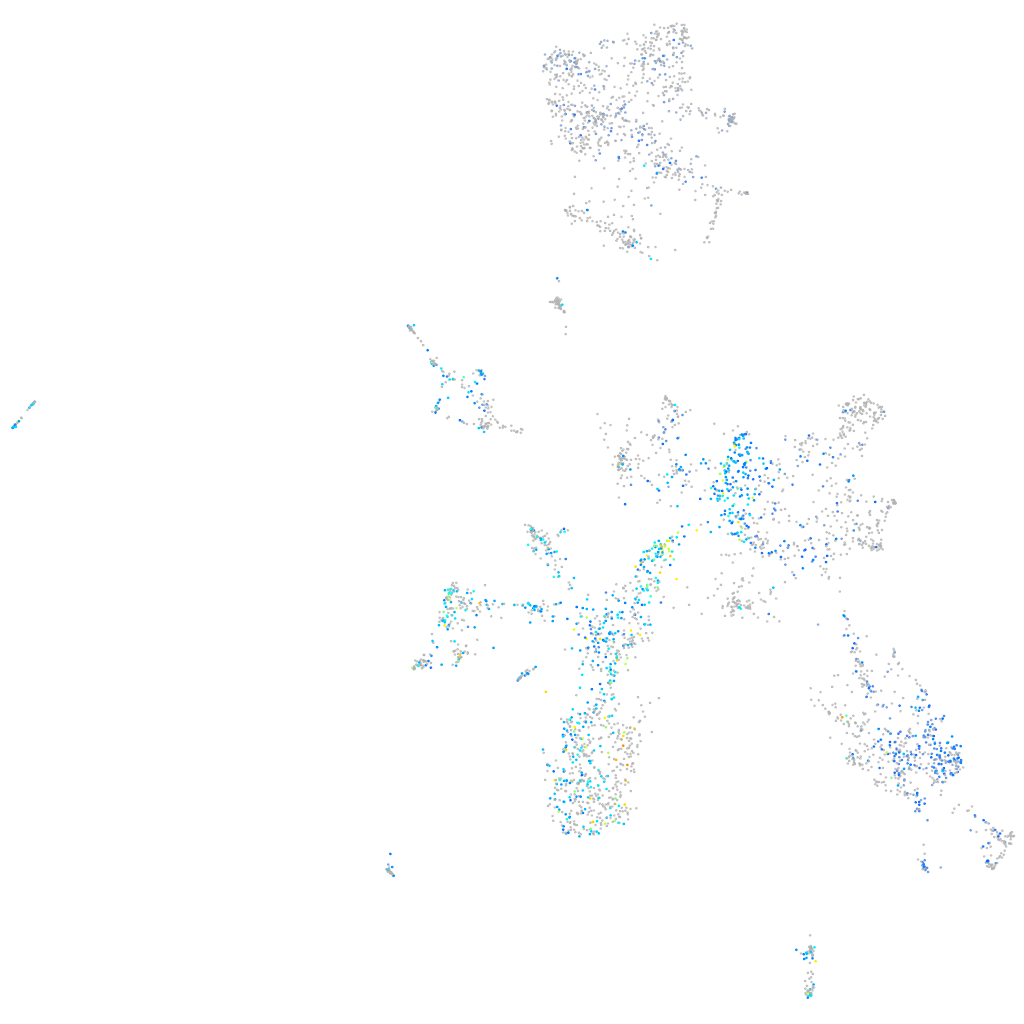
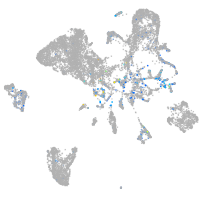
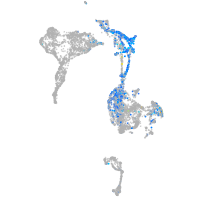
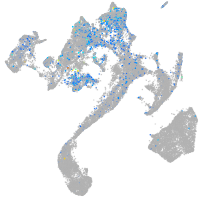
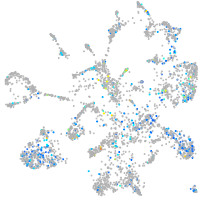
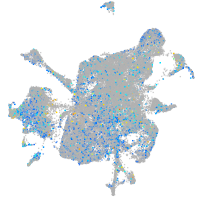
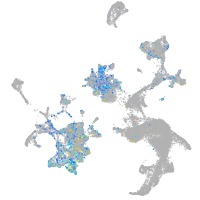
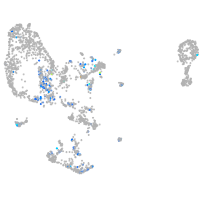
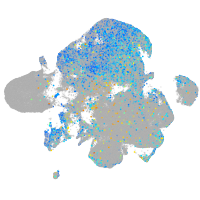
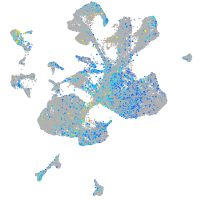
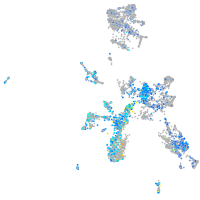
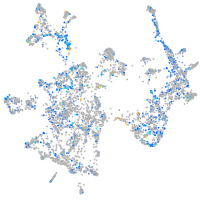
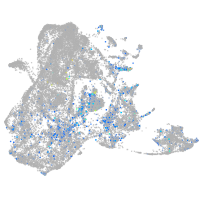
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| elavl3 | 0.405 | pfn1 | -0.283 |
| ebf3a | 0.350 | krt4 | -0.278 |
| insm1a | 0.346 | krt8 | -0.273 |
| neurod4 | 0.335 | lgals3b | -0.264 |
| ebf2 | 0.335 | krt18a.1 | -0.262 |
| insm1b | 0.329 | s100v2 | -0.261 |
| neurod1 | 0.327 | tmsb1 | -0.260 |
| myt1a | 0.309 | cfl1l | -0.258 |
| stmn1b | 0.306 | icn | -0.256 |
| sox11b | 0.305 | zgc:158343 | -0.246 |
| lhx2a | 0.301 | si:dkey-87o1.2 | -0.246 |
| CU467822.1 | 0.301 | zgc:92380 | -0.242 |
| tmeff1b | 0.289 | capns1a | -0.240 |
| uncx | 0.289 | rpz5 | -0.237 |
| sox11a | 0.287 | apoeb | -0.231 |
| epb41a | 0.284 | s100a10b | -0.231 |
| tuba1a | 0.276 | s100u | -0.231 |
| rtn1a | 0.273 | anxa4 | -0.230 |
| tmsb | 0.271 | eef1da | -0.230 |
| LOC100537342 | 0.270 | ahnak | -0.230 |
| tp53inp2 | 0.264 | sdc4 | -0.229 |
| ptmab | 0.262 | tagln2 | -0.228 |
| cxcr4b | 0.260 | s100v1 | -0.228 |
| dlb | 0.256 | sult6b1 | -0.226 |
| si:dkey-56m19.5 | 0.249 | capgb | -0.224 |
| si:ch211-193l2.6 | 0.249 | cldni | -0.222 |
| nhlh2 | 0.247 | egr1 | -0.222 |
| celf3a | 0.246 | zgc:153911 | -0.221 |
| hmgb3a | 0.241 | ier2a | -0.221 |
| gnb1b | 0.240 | GCA | -0.219 |
| nova2 | 0.238 | plecb | -0.218 |
| dla | 0.238 | crip1 | -0.217 |
| srrm4 | 0.235 | krt91 | -0.217 |
| ncam1a | 0.233 | krt97 | -0.216 |
| inka1a | 0.233 | cavin2b | -0.215 |