zinc finger protein 516
ZFIN 
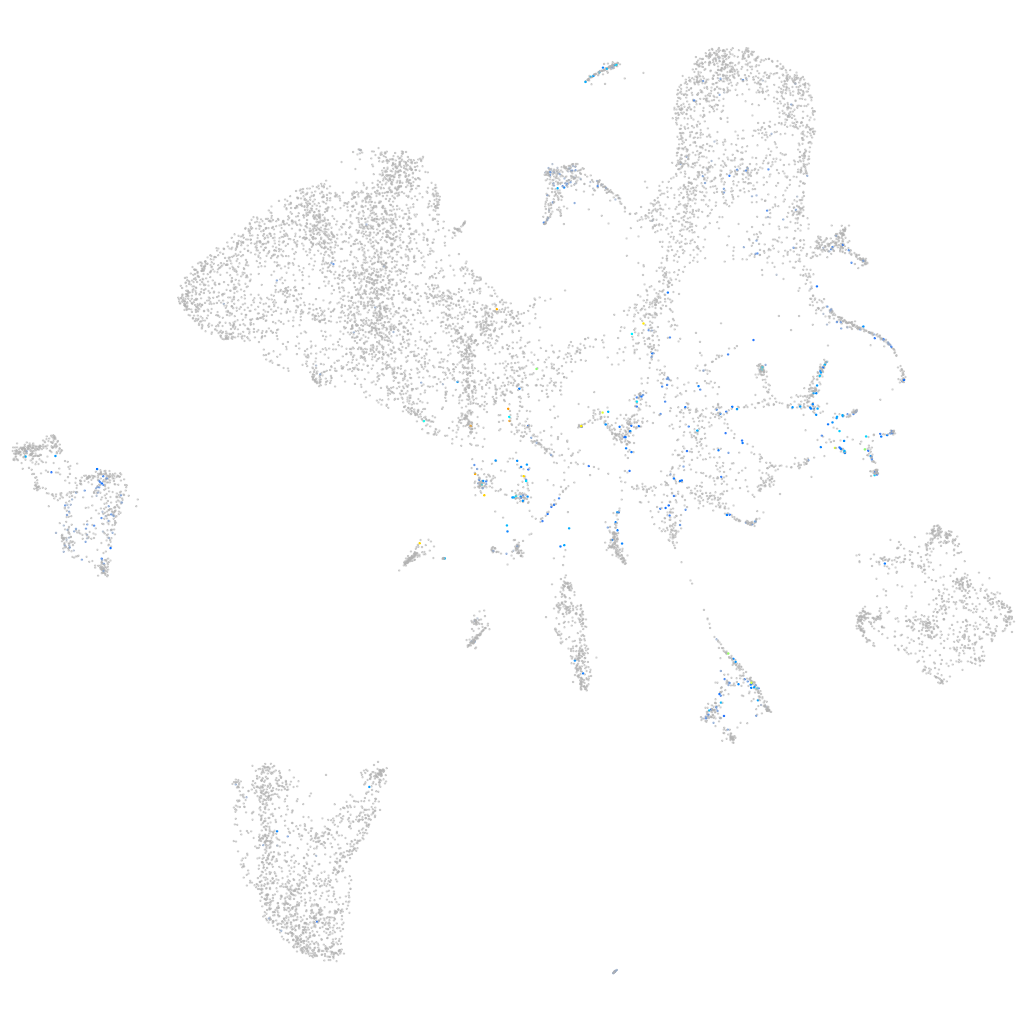
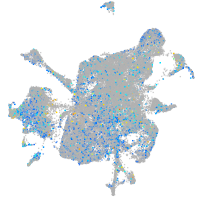
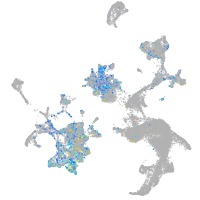
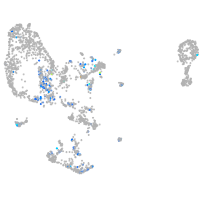
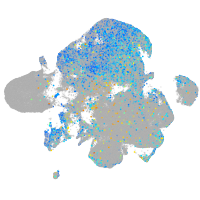
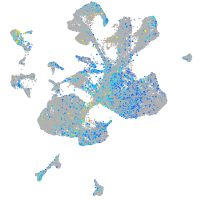
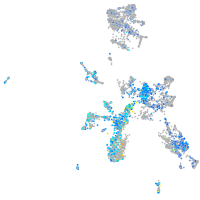
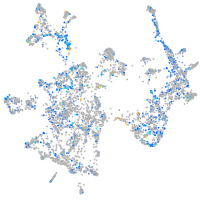
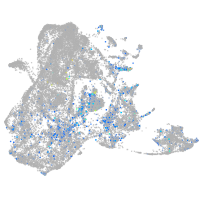
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| neurod1 | 0.199 | gapdh | -0.141 |
| insm1a | 0.183 | gamt | -0.134 |
| vat1 | 0.175 | gatm | -0.131 |
| pax6b | 0.173 | mat1a | -0.123 |
| scg3 | 0.173 | fbp1b | -0.121 |
| zgc:198371 | 0.171 | scp2a | -0.121 |
| gapdhs | 0.169 | apoa4b.1 | -0.119 |
| LOC101882396 | 0.166 | aldob | -0.118 |
| jpt1b | 0.166 | bhmt | -0.118 |
| msi1 | 0.164 | ahcy | -0.116 |
| socs5a | 0.162 | cx32.3 | -0.112 |
| si:ch1073-429i10.3.1 | 0.162 | aldh6a1 | -0.111 |
| rnasekb | 0.161 | glud1b | -0.110 |
| LOC110440001 | 0.159 | apoc2 | -0.109 |
| tox | 0.159 | agxtb | -0.108 |
| LOC100537384 | 0.158 | si:dkey-16p21.8 | -0.106 |
| si:dkey-153k10.9 | 0.157 | apoa1b | -0.106 |
| gpc1a | 0.154 | aqp12 | -0.104 |
| myt1b | 0.153 | gstt1a | -0.104 |
| CR383676.1 | 0.153 | agxta | -0.103 |
| scgn | 0.153 | pnp4b | -0.103 |
| lysmd2 | 0.153 | g6pca.2 | -0.102 |
| tusc3 | 0.152 | abat | -0.102 |
| mir7a-1 | 0.152 | eno3 | -0.101 |
| oaz1b | 0.151 | gcshb | -0.101 |
| gdi1 | 0.151 | apoc1 | -0.101 |
| ncam1a | 0.150 | ttc36 | -0.100 |
| plp1a | 0.149 | gnmt | -0.100 |
| calm1a | 0.149 | gstr | -0.099 |
| vamp2 | 0.148 | suclg1 | -0.099 |
| dpysl2b | 0.147 | tdo2a | -0.099 |
| XLOC-043912 | 0.147 | serpina1l | -0.099 |
| scg2a | 0.146 | gpx4a | -0.099 |
| csf2rb | 0.145 | phyhd1 | -0.098 |
| nkx2.2a | 0.143 | afp4 | -0.097 |