zinc finger protein 318
ZFIN 
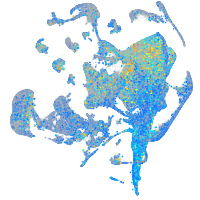
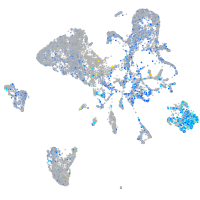
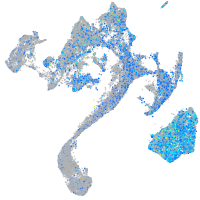
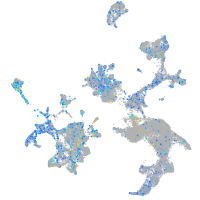
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| hmga1a | 0.124 | glula | -0.089 |
| cirbpa | 0.122 | slc1a2b | -0.084 |
| seta | 0.119 | atp1a1b | -0.081 |
| hnrnpaba | 0.118 | efhd1 | -0.080 |
| khdrbs1a | 0.115 | fabp7a | -0.079 |
| si:ch211-288g17.3 | 0.114 | cx43 | -0.076 |
| tubb2b | 0.111 | pvalb1 | -0.074 |
| si:ch211-222l21.1 | 0.111 | slc3a2a | -0.073 |
| hnrnpa0l | 0.111 | mt2 | -0.071 |
| h2afvb | 0.109 | acbd7 | -0.070 |
| marcksb | 0.108 | si:ch211-66e2.5 | -0.070 |
| hnrnpa0b | 0.108 | ppap2d | -0.070 |
| ptmab | 0.106 | ptn | -0.070 |
| hnrnpa1b | 0.105 | smox | -0.068 |
| nucks1a | 0.103 | cebpd | -0.067 |
| hnrnpabb | 0.103 | mdkb | -0.066 |
| ddx39ab | 0.102 | slc4a4a | -0.065 |
| h3f3a | 0.102 | hepacama | -0.065 |
| baz1b | 0.101 | cdo1 | -0.064 |
| hmgb2b | 0.101 | cd63 | -0.064 |
| cirbpb | 0.101 | pvalb2 | -0.064 |
| syncrip | 0.100 | gpr37l1b | -0.063 |
| cbx3a | 0.099 | qki2 | -0.063 |
| snrpd1 | 0.097 | cox4i2 | -0.063 |
| setb | 0.097 | apoa2 | -0.062 |
| snrpb | 0.096 | anxa13 | -0.060 |
| eef2b | 0.095 | rpz5 | -0.060 |
| hdac1 | 0.095 | mfge8a | -0.060 |
| ppm1g | 0.094 | eno1b | -0.060 |
| hsp90ab1 | 0.092 | slc6a1b | -0.059 |
| atrx | 0.092 | dap1b | -0.058 |
| snrpe | 0.091 | itm2ba | -0.057 |
| hnrnpub | 0.091 | ppdpfb | -0.057 |
| snrpd2 | 0.090 | chp2 | -0.057 |
| taf15 | 0.090 | cspg5b | -0.056 |