"zinc finger, matrin-type 4a"
ZFIN 
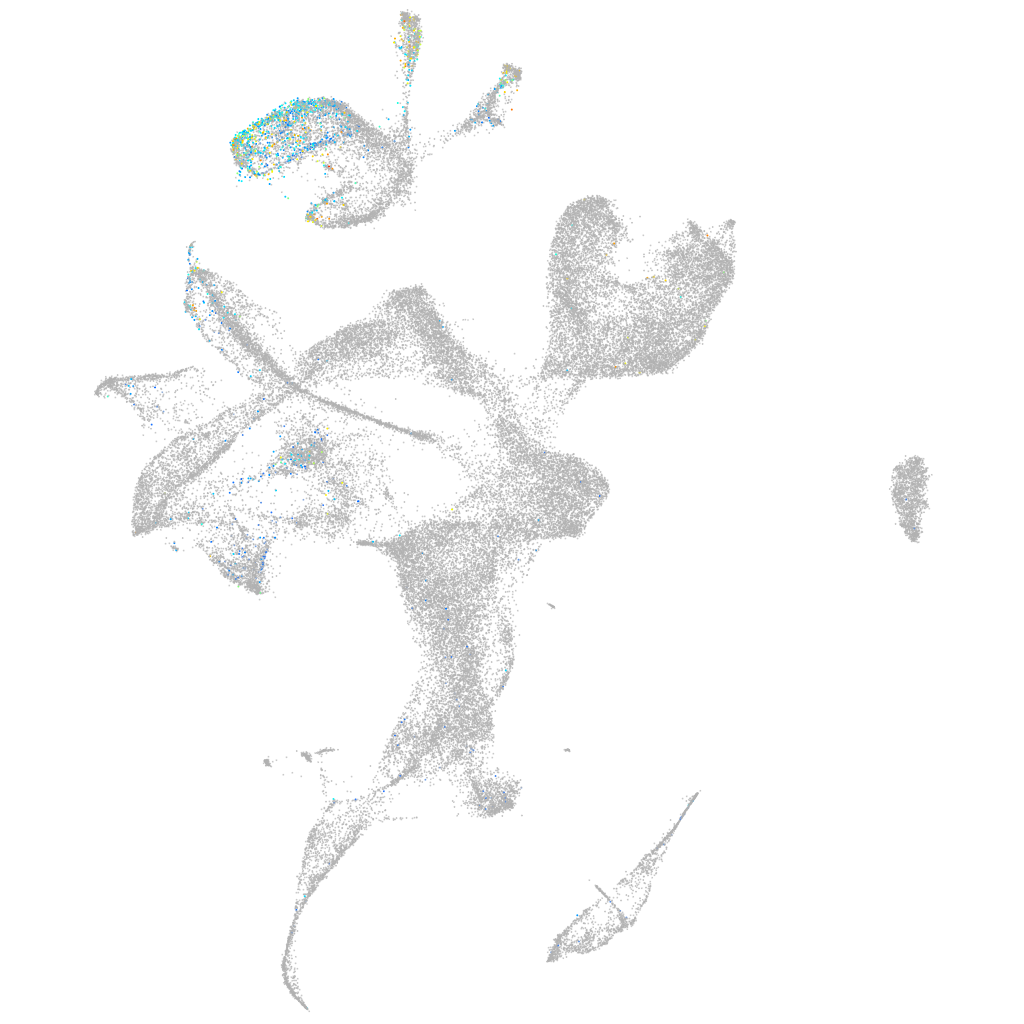
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| syt1a | 0.323 | hmgb2a | -0.176 |
| slc32a1 | 0.316 | ahcy | -0.094 |
| slc6a1a | 0.304 | hmgb2b | -0.092 |
| stxbp1a | 0.292 | mdka | -0.092 |
| tfap2a | 0.288 | fabp7a | -0.090 |
| sv2a | 0.280 | stmn1a | -0.089 |
| stx1b | 0.276 | msi1 | -0.087 |
| tfap2b | 0.267 | hmga1a | -0.087 |
| tfap2e | 0.265 | pcna | -0.086 |
| LOC101886114 | 0.257 | msna | -0.082 |
| ywhag2 | 0.252 | rrm1 | -0.080 |
| cabp1b | 0.251 | dut | -0.080 |
| lrtm1 | 0.249 | rplp1 | -0.079 |
| cplx3b | 0.241 | mki67 | -0.078 |
| slc6a1b | 0.237 | lbr | -0.078 |
| mir181b-3 | 0.234 | nutf2l | -0.078 |
| gad2 | 0.232 | tuba8l4 | -0.077 |
| pax10 | 0.232 | rplp2l | -0.077 |
| si:ch73-119p20.1 | 0.231 | tuba8l | -0.077 |
| fosl2 | 0.230 | rpsa | -0.077 |
| dtnbp1b | 0.230 | rpl12 | -0.076 |
| eno2 | 0.229 | rps20 | -0.076 |
| c1ql1a | 0.225 | ccnd1 | -0.076 |
| sncb | 0.223 | neurod4 | -0.075 |
| meis2a | 0.219 | rps19 | -0.075 |
| syt6a | 0.218 | her15.1 | -0.075 |
| tkta | 0.216 | chaf1a | -0.074 |
| slc35g2b | 0.216 | tspan7 | -0.074 |
| celf5a | 0.214 | hes6 | -0.073 |
| syn2b | 0.214 | mcm7 | -0.073 |
| nrxn1a | 0.214 | si:dkey-151g10.6 | -0.073 |
| tfap2c | 0.211 | tpt1 | -0.072 |
| znf385a | 0.207 | cx43.4 | -0.072 |
| syn2a | 0.206 | ccna2 | -0.072 |
| gad1b | 0.204 | crx | -0.072 |