zgc:77151
ZFIN 
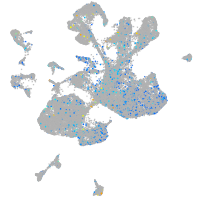
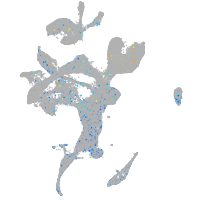
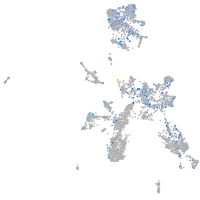
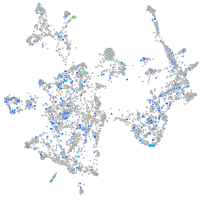
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| cx43.4 | 0.068 | gapdhs | -0.045 |
| khdrbs1a | 0.065 | atp6v0cb | -0.045 |
| hmga1a | 0.064 | sncb | -0.043 |
| sb:cb81 | 0.063 | ywhag2 | -0.042 |
| msna | 0.060 | atp6ap2 | -0.039 |
| id1 | 0.059 | rnasekb | -0.039 |
| chd7 | 0.059 | sncgb | -0.038 |
| seta | 0.059 | tpi1b | -0.038 |
| lfng | 0.058 | snap25a | -0.038 |
| hnrnpa0b | 0.058 | eno2 | -0.037 |
| si:ch1073-80i24.3 | 0.058 | atpv0e2 | -0.037 |
| notch3 | 0.057 | pvalb1 | -0.037 |
| top1l | 0.057 | elavl4 | -0.037 |
| mki67 | 0.056 | zgc:65894 | -0.036 |
| hnrnpa1b | 0.056 | rtn1b | -0.035 |
| tspan7 | 0.056 | gng3 | -0.035 |
| smc1al | 0.055 | pvalb2 | -0.035 |
| cirbpb | 0.055 | sypb | -0.035 |
| ddx39ab | 0.055 | calm1b | -0.035 |
| sox19a | 0.055 | ndufa4 | -0.035 |
| hnrnpub | 0.054 | tmsb2 | -0.035 |
| ccnd1 | 0.054 | stmn2a | -0.035 |
| akap12b | 0.054 | map1aa | -0.034 |
| syncrip | 0.054 | calm1a | -0.034 |
| si:ch211-288g17.3 | 0.054 | rab6bb | -0.034 |
| hdac1 | 0.054 | gpm6ab | -0.034 |
| cirbpa | 0.054 | basp1 | -0.033 |
| taf15 | 0.053 | slc6a1a | -0.033 |
| fbxo5 | 0.053 | stxbp1a | -0.033 |
| XLOC-003689 | 0.053 | si:ch73-119p20.1 | -0.032 |
| hmgb2b | 0.053 | syn2a | -0.032 |
| pou5f3 | 0.053 | nptna | -0.032 |
| ctsla | 0.053 | vamp2 | -0.032 |
| sox3 | 0.053 | atp6v1g1 | -0.032 |
| XLOC-003690 | 0.053 | stx1b | -0.032 |