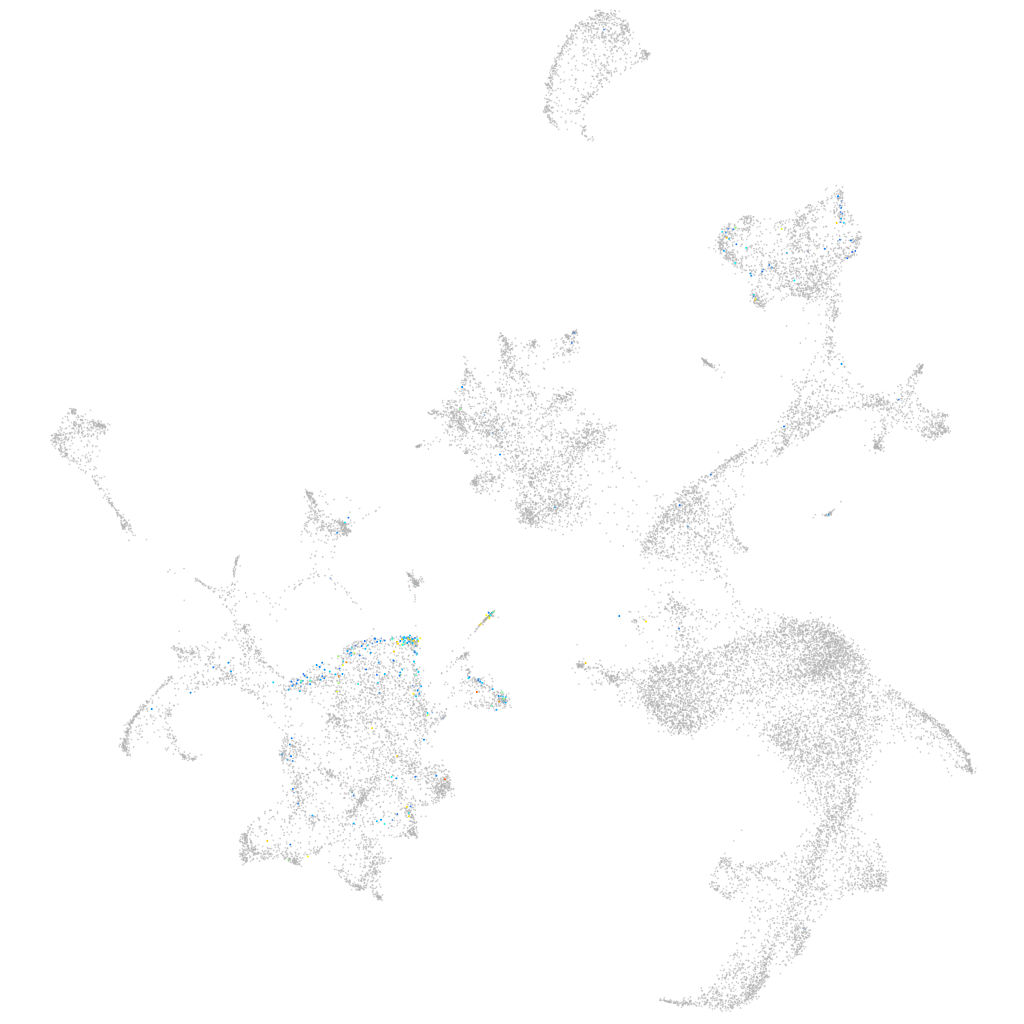
zgc:174356
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| mrc1a | 0.267 | hbae1.1 | -0.098 |
| lyve1b | 0.261 | hbae1.3 | -0.092 |
| stab2 | 0.244 | hbae3 | -0.092 |
| si:dkey-28n18.9 | 0.241 | hbbe2 | -0.091 |
| stab1 | 0.240 | hbbe1.1 | -0.090 |
| dab2 | 0.237 | hbbe1.3 | -0.090 |
| lgmn | 0.236 | cahz | -0.087 |
| npl | 0.233 | hemgn | -0.081 |
| hyal2b | 0.215 | hbbe1.2 | -0.079 |
| hexb | 0.215 | fth1a | -0.078 |
| osr2 | 0.214 | alas2 | -0.077 |
| lamp2 | 0.212 | blvrb | -0.076 |
| cpn1 | 0.210 | si:ch211-250g4.3 | -0.072 |
| ctsla | 0.208 | slc4a1a | -0.072 |
| tpcn3 | 0.205 | nt5c2l1 | -0.070 |
| glula | 0.202 | tspo | -0.070 |
| fuca1.1 | 0.199 | zgc:163057 | -0.067 |
| zgc:110239 | 0.198 | epb41b | -0.067 |
| tpte | 0.196 | si:ch211-207c6.2 | -0.063 |
| tmem106a | 0.195 | prdx2 | -0.062 |
| timp2b | 0.187 | creg1 | -0.061 |
| gnpda1 | 0.187 | nmt1b | -0.060 |
| cpvl | 0.183 | plac8l1 | -0.059 |
| ctsh | 0.181 | tmod4 | -0.059 |
| ap1b1 | 0.181 | zgc:56095 | -0.057 |
| gpr182 | 0.179 | gpx1a | -0.056 |
| snx2 | 0.178 | anp32b | -0.054 |
| fuca2 | 0.177 | hbae5 | -0.054 |
| dpp7 | 0.175 | tfr1a | -0.053 |
| pttg1ipb | 0.174 | sptb | -0.053 |
| b3gnt7 | 0.172 | znfl2a | -0.052 |
| zgc:110591 | 0.171 | rfesd | -0.052 |
| ctgfa | 0.170 | uros | -0.050 |
| snx8a | 0.169 | hdr | -0.050 |
| hvcn1 | 0.168 | hbbe3 | -0.050 |