zgc:174164
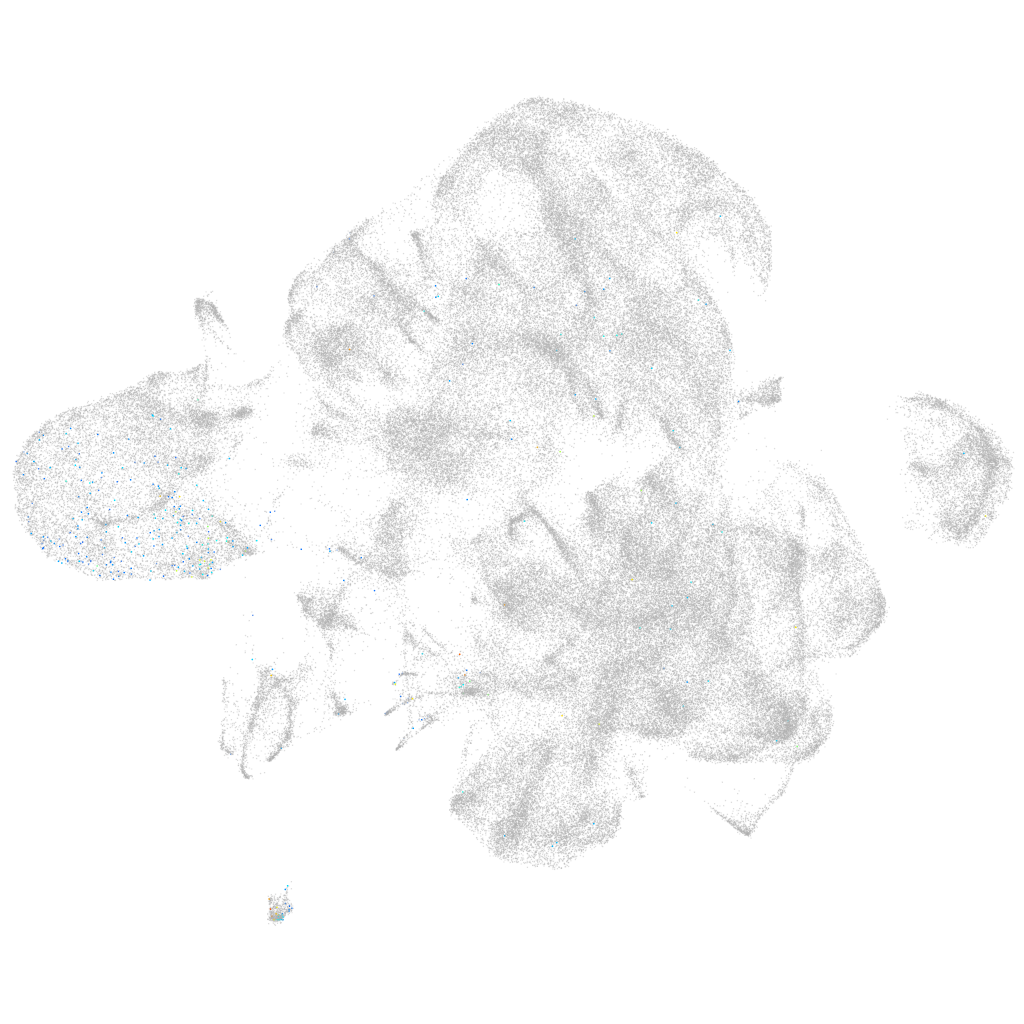
ZFIN 
Other cell groups


all cells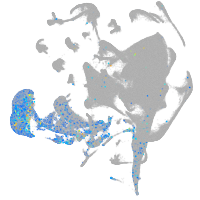 |
blastomeres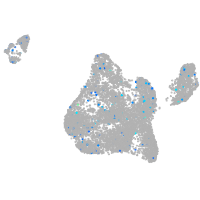 |
PGCs |
endoderm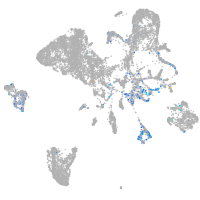 |
axial mesoderm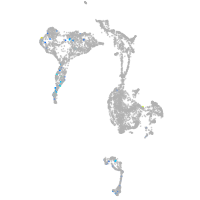 |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme 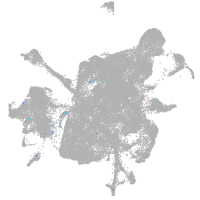 |
hematopoietic / vasculature 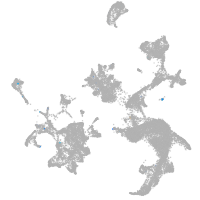 |
pronephros  |
neural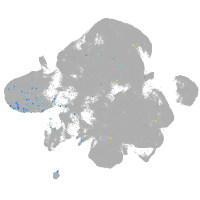 |
spinal cord / glia |
eye |
taste / olfactory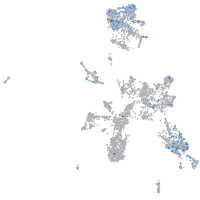 |
otic / lateral line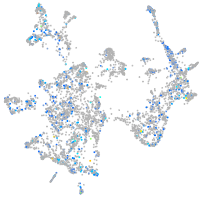 |
epidermis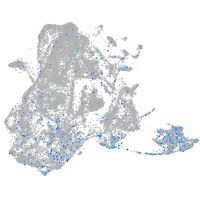 |
periderm |
fin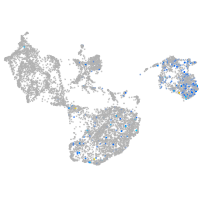 |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| si:dkey-37o8.1 | 0.103 | rpl37 | -0.055 |
| LOC108181504 | 0.092 | rps10 | -0.054 |
| dlx3b | 0.091 | zgc:114188 | -0.052 |
| epcam | 0.090 | h3f3a | -0.050 |
| bspry | 0.087 | hnrnpa0l | -0.048 |
| apoc1 | 0.084 | fabp3 | -0.048 |
| psmb8a | 0.084 | tmsb4x | -0.048 |
| tfap2c | 0.080 | ptmaa | -0.045 |
| XLOC-005215 | 0.079 | marcksl1a | -0.042 |
| foxi1 | 0.078 | tuba1c | -0.040 |
| apoeb | 0.076 | nova2 | -0.038 |
| nipal4 | 0.076 | rps17 | -0.038 |
| CR376800.1 | 0.076 | ppiab | -0.033 |
| f11r.1 | 0.076 | si:ch1073-429i10.3.1 | -0.033 |
| si:ch211-256e16.8 | 0.074 | elavl3 | -0.032 |
| zgc:112038 | 0.073 | cct2 | -0.031 |
| tagln2 | 0.073 | gpm6aa | -0.031 |
| cldnh | 0.072 | nme2b.1 | -0.030 |
| cdh1 | 0.069 | eef1g | -0.030 |
| arhgef19 | 0.068 | rtn1a | -0.029 |
| tp63 | 0.068 | idh2 | -0.028 |
| b4galnt3b | 0.067 | stmn1b | -0.027 |
| BX294388.1 | 0.067 | eef1b2 | -0.027 |
| ved | 0.065 | tubb2b | -0.027 |
| plcd1b | 0.065 | celf2 | -0.027 |
| cldn7b | 0.065 | rpl19 | -0.027 |
| si:ch211-152c2.3 | 0.064 | h3f3c | -0.027 |
| si:ch211-130m23.2 | 0.064 | CR383676.1 | -0.027 |
| CR381686.2 | 0.063 | fam168a | -0.026 |
| dbnla | 0.062 | tubb4b | -0.026 |
| spint2 | 0.062 | gpm6ab | -0.026 |
| cldnb | 0.062 | tmsb | -0.026 |
| tor1l3 | 0.061 | zgc:158463 | -0.026 |
| prdm1a | 0.061 | rpl9 | -0.025 |
| hepacam2 | 0.061 | rpl24 | -0.025 |