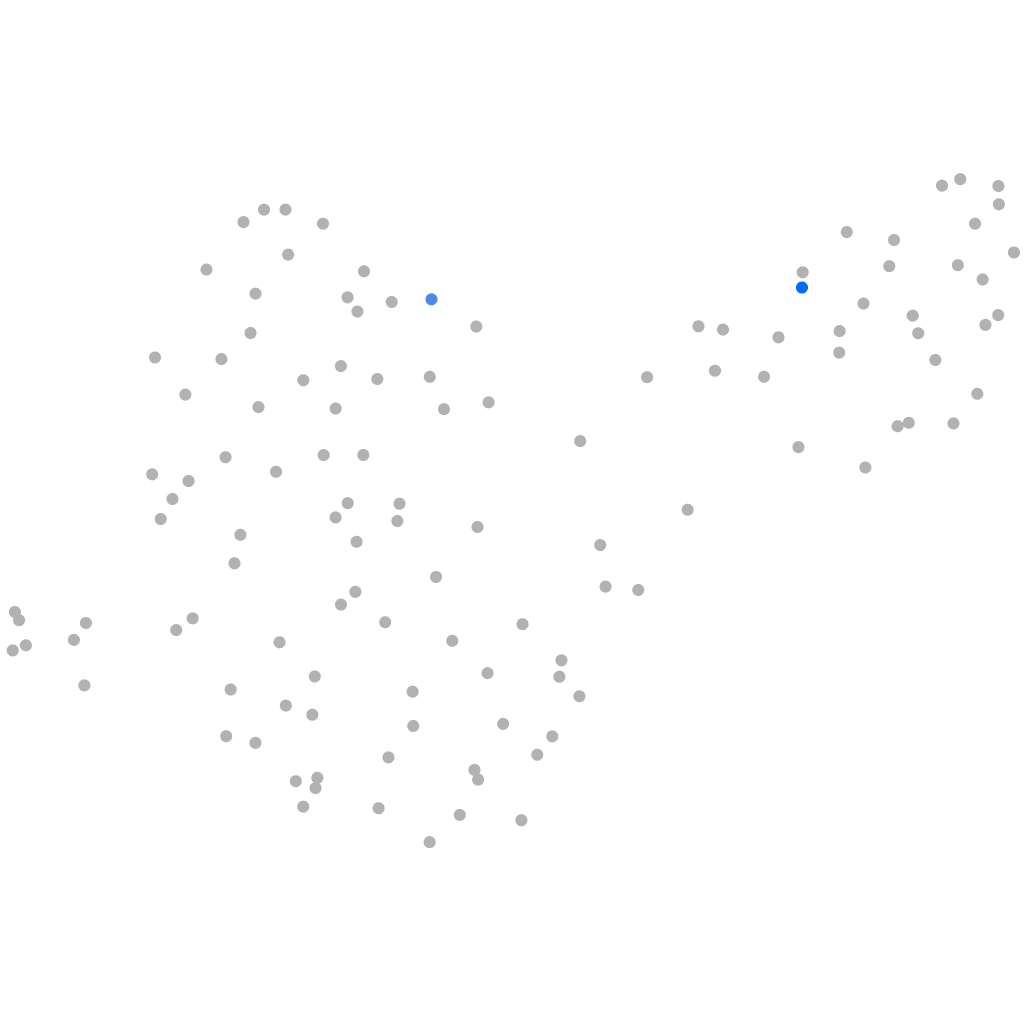
zgc:158432
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| bsnb | 1.000 | hnrnpaba | -0.197 |
| faxcb | 1.000 | snrpa | -0.196 |
| lrp1ba | 1.000 | nucks1a | -0.168 |
| nptx2b | 1.000 | brd3a | -0.155 |
| sema7a | 1.000 | top1l | -0.153 |
| sv2c | 1.000 | gpkow | -0.150 |
| tmem150c | 0.842 | smarca4a | -0.148 |
| cldn15lb | 0.842 | srrt | -0.147 |
| nr1i2 | 0.842 | akap12b | -0.145 |
| nrxn3a | 0.842 | cfl1 | -0.144 |
| opn8b | 0.842 | sltm | -0.141 |
| XLOC-019725 | 0.842 | ptmab | -0.139 |
| AL935300.3 | 0.842 | tpx2 | -0.138 |
| ank2a | 0.842 | ccng2 | -0.138 |
| arhgef33 | 0.842 | srsf4 | -0.135 |
| BX005073.3 | 0.842 | hmgb1a | -0.134 |
| chrm4a | 0.842 | g3bp1 | -0.133 |
| CT027702.1 | 0.842 | tcea1 | -0.130 |
| CU024870.1 | 0.842 | nucks1b | -0.129 |
| CU457778.1 | 0.842 | atp1a1a.1 | -0.129 |
| CU693446.1 | 0.842 | rbm7 | -0.127 |
| dpf3 | 0.842 | spcs2 | -0.127 |
| ERG28 | 0.842 | eif4ebp3l | -0.126 |
| ern2 | 0.842 | nop56 | -0.125 |
| espnla | 0.842 | apoeb | -0.124 |
| fam19a5b | 0.842 | UTP14C | -0.124 |
| FO704822.1 | 0.842 | hnrnpa0a | -0.123 |
| glipr1b | 0.842 | ppig | -0.122 |
| klhdc7a | 0.842 | marcksb | -0.121 |
| klhl23 | 0.842 | cbx3a | -0.120 |
| krt1-19d | 0.842 | anp32a | -0.120 |
| LO017656.1 | 0.842 | kif23 | -0.117 |
| LOC100537263 | 0.842 | pdcd10a | -0.117 |
| LOC101882330 | 0.842 | cnn3b | -0.117 |
| LOC108190601 | 0.842 | nelfe | -0.117 |