zgc:158320
ZFIN 
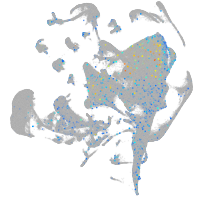
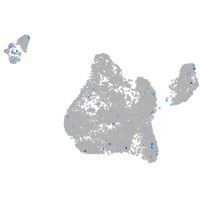
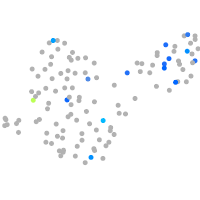
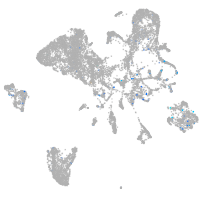
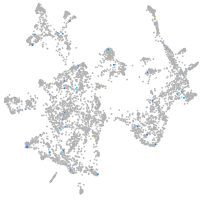
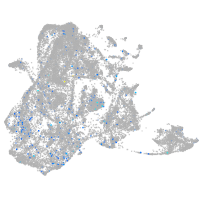
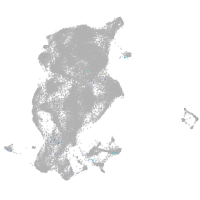
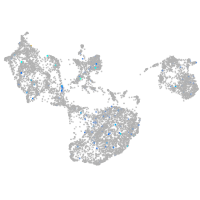
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| neto2b | 0.111 | ttn.1 | -0.063 |
| LOC103911320 | 0.100 | ttn.2 | -0.061 |
| CT956055.1 | 0.094 | aldoab | -0.056 |
| CR391991.4 | 0.084 | atp2a1 | -0.054 |
| ndufa4l2a | 0.081 | ak1 | -0.054 |
| tmsb4x | 0.078 | mybphb | -0.053 |
| marcksl1b | 0.075 | tnnc2 | -0.053 |
| RNF208 | 0.074 | tmem38a | -0.053 |
| mfrp | 0.070 | klhl41b | -0.053 |
| LO018513.1 | 0.069 | ckmb | -0.053 |
| pdia3 | 0.068 | actc1b | -0.052 |
| tm4sf5 | 0.066 | hsp90aa1.1 | -0.052 |
| kcne4 | 0.065 | desma | -0.052 |
| trim35-25 | 0.065 | srl | -0.052 |
| ctsla | 0.064 | tpma | -0.051 |
| foxf2b | 0.064 | CABZ01078594.1 | -0.051 |
| marcksl1a | 0.063 | ldb3a | -0.050 |
| CR377229.1 | 0.063 | acta1a | -0.050 |
| BX000434.1 | 0.062 | cav3 | -0.050 |
| actb1 | 0.062 | pabpc4 | -0.050 |
| pde5ab | 0.062 | ckma | -0.049 |
| gprin1 | 0.062 | neb | -0.049 |
| actb2 | 0.062 | zgc:101853 | -0.049 |
| si:ch211-168d23.3 | 0.062 | actn3b | -0.048 |
| lasp1 | 0.061 | txlnbb | -0.048 |
| fthl27 | 0.061 | CABZ01072309.1 | -0.048 |
| LOC101884484 | 0.061 | cycsb | -0.048 |
| BX511079.2 | 0.061 | zgc:92429 | -0.048 |
| tpm4a | 0.061 | actc1a | -0.047 |
| krt18b | 0.061 | rbfox1l | -0.047 |
| pmp22a | 0.060 | actn3a | -0.047 |
| si:dkeyp-75h12.5 | 0.060 | acta1b | -0.047 |
| cfl1 | 0.060 | tnni2b.1 | -0.047 |
| ywhaz | 0.059 | gapdh | -0.047 |
| ptmaa | 0.059 | casq2 | -0.046 |