zgc:158263
ZFIN 
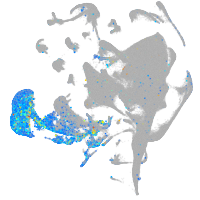
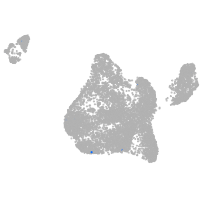
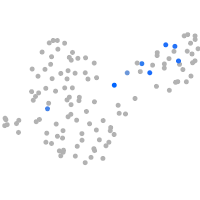
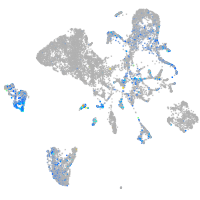
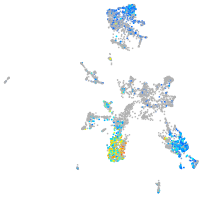
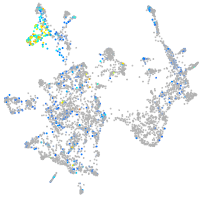
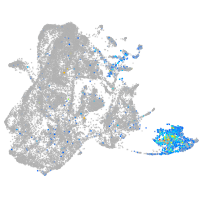
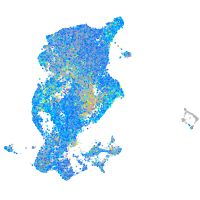
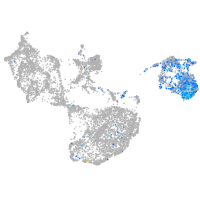
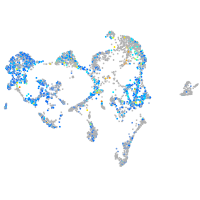
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| suclg1 | 0.259 | marcksb | -0.212 |
| ldhba | 0.256 | si:ch73-281n10.2 | -0.212 |
| atp5mc3b | 0.255 | nucks1a | -0.212 |
| mdh1aa | 0.253 | hspb1 | -0.210 |
| atp5f1b | 0.253 | si:ch73-1a9.3 | -0.209 |
| atp5mc1 | 0.252 | hnrnpaba | -0.207 |
| si:ch211-139a5.9 | 0.251 | marcksl1b | -0.206 |
| cdaa | 0.248 | wu:fb97g03 | -0.205 |
| ca2 | 0.247 | apoeb | -0.198 |
| atp5fa1 | 0.246 | ptmab | -0.195 |
| eno3 | 0.242 | hmgb1b | -0.194 |
| glud1b | 0.242 | hmgb2b | -0.191 |
| cox7a2a | 0.241 | hnrnpabb | -0.190 |
| sod2 | 0.240 | cx43.4 | -0.188 |
| COX5B | 0.239 | apoc1 | -0.185 |
| atp1b1a | 0.238 | syncrip | -0.185 |
| ndrg1a | 0.237 | anp32a | -0.181 |
| cox6a1 | 0.235 | setb | -0.180 |
| atp5l | 0.235 | hmgn6 | -0.179 |
| cdh17 | 0.235 | cbx3a | -0.179 |
| atp5f1d | 0.232 | tubb2b | -0.179 |
| cox5ab | 0.231 | seta | -0.179 |
| cox6a2 | 0.231 | h2afvb | -0.176 |
| mt-nd2 | 0.230 | msna | -0.175 |
| ndufa10 | 0.229 | anp32b | -0.174 |
| si:ch1073-443f11.2 | 0.228 | anp32e | -0.172 |
| sdha | 0.227 | akap12b | -0.171 |
| ppifb | 0.226 | ncl | -0.171 |
| atp5meb | 0.226 | hmgb2a | -0.169 |
| mt-nd4 | 0.226 | top1l | -0.166 |
| ndufb3 | 0.225 | snrnp70 | -0.166 |
| bin2a | 0.223 | acin1a | -0.166 |
| atp5pd | 0.223 | hnrnpa0b | -0.166 |
| cox8a | 0.222 | dkc1 | -0.166 |
| eps8l3a | 0.222 | nono | -0.166 |