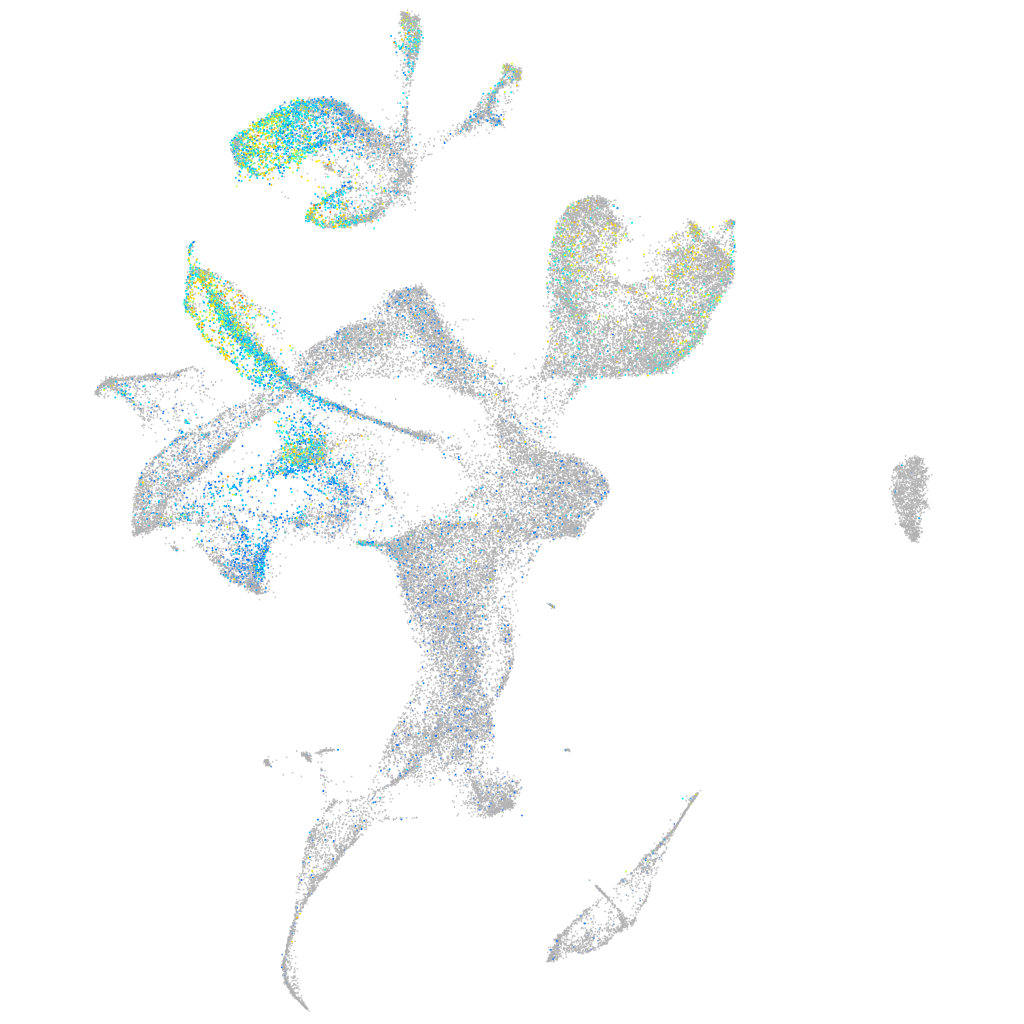zgc:153426
ZFIN 
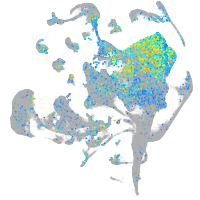
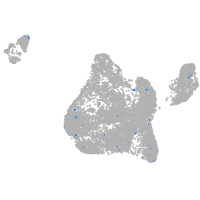
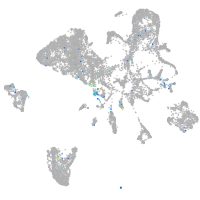
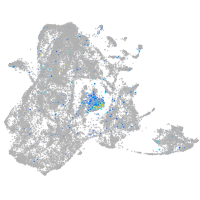
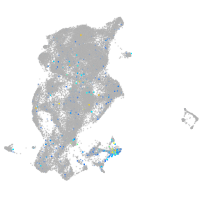
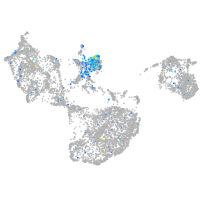
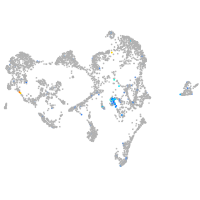
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| rtn1b | 0.469 | hmgb2a | -0.313 |
| ywhag2 | 0.451 | stmn1a | -0.185 |
| gng3 | 0.445 | pcna | -0.184 |
| zgc:65894 | 0.443 | ahcy | -0.180 |
| eno2 | 0.433 | mdka | -0.180 |
| stmn2a | 0.432 | hmga1a | -0.177 |
| stxbp1a | 0.430 | rrm1 | -0.172 |
| stx1b | 0.414 | nutf2l | -0.169 |
| elavl3 | 0.402 | lbr | -0.167 |
| sncb | 0.399 | mki67 | -0.167 |
| ppp1r14ba | 0.396 | ccnd1 | -0.166 |
| snap25a | 0.383 | chaf1a | -0.164 |
| stmn1b | 0.375 | fabp7a | -0.164 |
| syt1a | 0.363 | msi1 | -0.164 |
| mllt11 | 0.363 | dut | -0.163 |
| sv2a | 0.360 | msna | -0.162 |
| id4 | 0.359 | mcm7 | -0.161 |
| tuba1c | 0.357 | selenoh | -0.159 |
| maptb | 0.353 | tuba8l | -0.159 |
| elavl4 | 0.350 | ccna2 | -0.153 |
| syt2a | 0.349 | hmgb2b | -0.152 |
| rbfox1 | 0.349 | dek | -0.151 |
| sprn | 0.348 | banf1 | -0.151 |
| cplx2l | 0.348 | lig1 | -0.148 |
| tuba2 | 0.344 | COX7A2 (1 of many) | -0.147 |
| gng2 | 0.342 | eef1da | -0.147 |
| si:ch211-129p13.1 | 0.340 | her15.1 | -0.146 |
| gabrb4 | 0.334 | cks1b | -0.145 |
| st8sia5 | 0.333 | rpa3 | -0.143 |
| rbpms2a | 0.332 | mcm6 | -0.143 |
| gnao1a | 0.331 | fen1 | -0.142 |
| gap43 | 0.330 | cx43.4 | -0.141 |
| si:ch73-119p20.1 | 0.328 | rrm2 | -0.138 |
| carmil2 | 0.325 | dlgap5 | -0.138 |
| islr2 | 0.324 | CABZ01005379.1 | -0.138 |