zgc:153039
ZFIN 
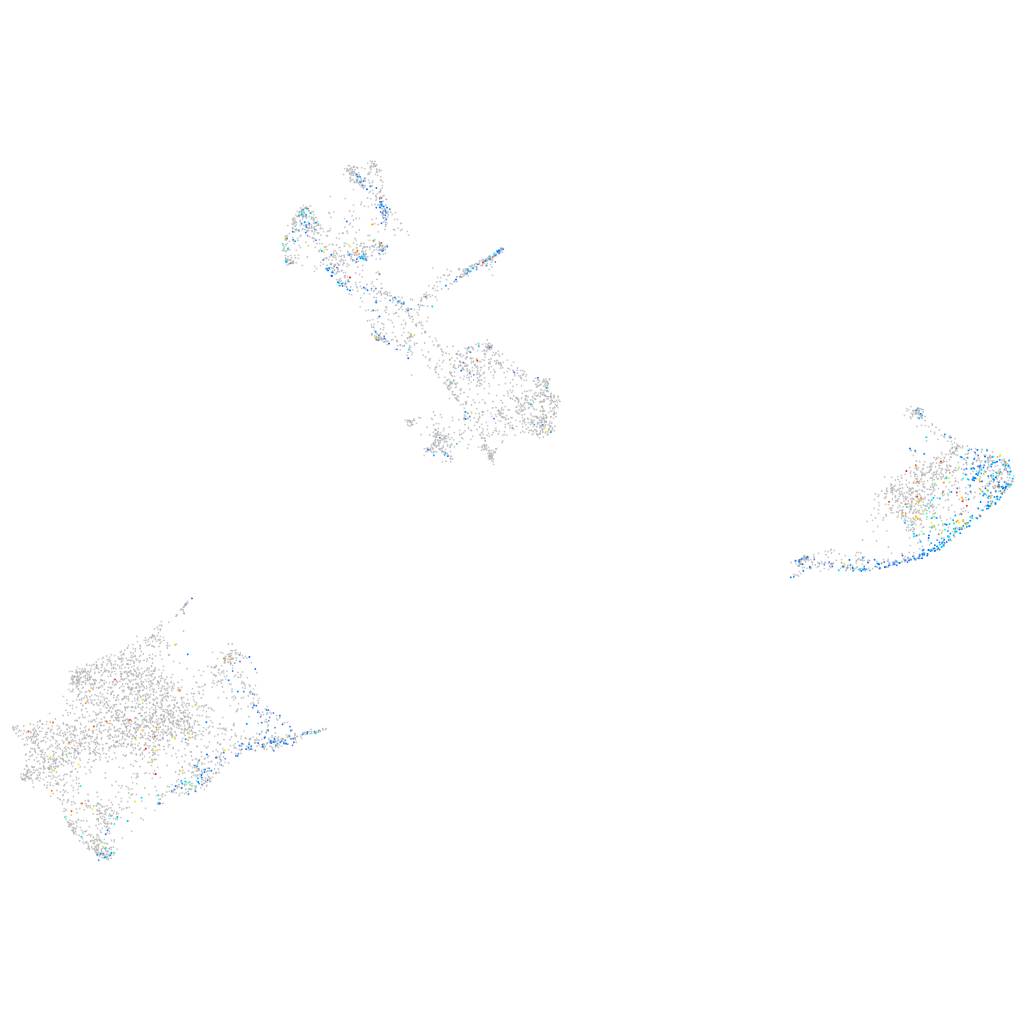
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| tyrp1a | 0.253 | CABZ01021592.1 | -0.159 |
| dct | 0.244 | si:dkey-251i10.2 | -0.128 |
| kita | 0.243 | uraha | -0.098 |
| tyr | 0.242 | paics | -0.094 |
| slc24a5 | 0.239 | tmem130 | -0.093 |
| SPAG9 | 0.239 | aldob | -0.085 |
| oca2 | 0.233 | si:ch211-222l21.1 | -0.085 |
| tyrp1b | 0.226 | si:ch73-1a9.3 | -0.084 |
| pmela | 0.224 | si:ch211-251b21.1 | -0.084 |
| slc7a5 | 0.223 | ptmaa | -0.080 |
| aadac | 0.221 | mdh1aa | -0.080 |
| slc45a2 | 0.219 | hmgb1b | -0.079 |
| slc37a2 | 0.217 | gpm6aa | -0.079 |
| slc24a4a | 0.214 | sult1st1 | -0.079 |
| slc29a3 | 0.211 | cyb5a | -0.078 |
| mtbl | 0.211 | aox5 | -0.076 |
| si:ch73-389b16.1 | 0.210 | nova2 | -0.076 |
| atp6v0ca | 0.210 | bco1 | -0.075 |
| lamp1a | 0.205 | si:ch73-281n10.2 | -0.073 |
| slc30a8 | 0.205 | hmgn2 | -0.072 |
| atp11a | 0.203 | tuba1c | -0.071 |
| psap | 0.202 | tmsb | -0.070 |
| atp6v0a2b | 0.201 | pvalb2 | -0.065 |
| mchr2 | 0.200 | CU467822.1 | -0.065 |
| kcnj13 | 0.194 | hbbe1.3 | -0.063 |
| zgc:91968 | 0.194 | stmn1b | -0.063 |
| bace2 | 0.192 | tox | -0.063 |
| tspan10 | 0.192 | prdx1 | -0.063 |
| rab38 | 0.190 | elavl3 | -0.062 |
| slc3a2a | 0.189 | zgc:153031 | -0.062 |
| birc7 | 0.188 | hbae3 | -0.060 |
| tspan36 | 0.188 | aqp3a | -0.060 |
| pttg1ipb | 0.186 | gch2 | -0.059 |
| CDK18 | 0.184 | oacyl | -0.059 |
| prkar1b | 0.183 | hmgb3a | -0.058 |