zgc:110239
ZFIN 
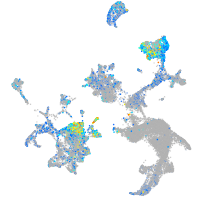
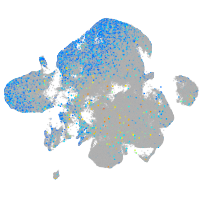
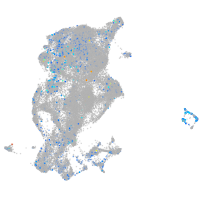
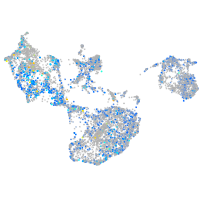
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| ctsla | 0.103 | fgfbp2b | -0.063 |
| mfap2 | 0.085 | rps29 | -0.051 |
| dcn | 0.082 | acana | -0.045 |
| pmp22a | 0.079 | wwp2 | -0.044 |
| marcksl1a | 0.073 | ppp1r1c | -0.042 |
| colec12 | 0.072 | ptmaa | -0.041 |
| lxn | 0.070 | rpl38 | -0.040 |
| sdc4 | 0.070 | emilin3a | -0.039 |
| pcolcea | 0.069 | mia | -0.036 |
| sparc | 0.069 | rpl29 | -0.036 |
| ctsd | 0.069 | loxl4 | -0.036 |
| cd63 | 0.068 | snorc | -0.035 |
| lum | 0.067 | si:dkey-19b23.8 | -0.034 |
| serpinf1 | 0.067 | rpl39 | -0.033 |
| angptl4 | 0.067 | rps21 | -0.033 |
| bsg | 0.066 | mob2b | -0.033 |
| pdgfrl | 0.066 | matn1 | -0.033 |
| fstl1a | 0.066 | rpl37 | -0.033 |
| pon3.2 | 0.066 | ptgdsa | -0.032 |
| col1a2 | 0.066 | col11a2 | -0.032 |
| wls | 0.065 | elavl3 | -0.031 |
| pcna | 0.065 | sox9a | -0.031 |
| twist1a | 0.065 | psmd2 | -0.031 |
| si:ch211-286o17.1 | 0.064 | ucmab | -0.030 |
| sept2 | 0.064 | hapln1b | -0.030 |
| akap12b | 0.063 | acanb | -0.030 |
| col5a1 | 0.062 | gpm6aa | -0.029 |
| cnn3a | 0.062 | abtb2a | -0.029 |
| cyr61 | 0.061 | hapln1a | -0.029 |
| zgc:110540 | 0.061 | si:ch211-156j16.1 | -0.029 |
| dhfr | 0.060 | klf2b | -0.028 |
| tuba8l | 0.060 | rtn1a | -0.028 |
| tuba8l4 | 0.060 | tmem178b | -0.028 |
| prdx4 | 0.060 | id4 | -0.027 |
| ctsba | 0.060 | adgrl3.1 | -0.027 |