zgc:110216
ZFIN 
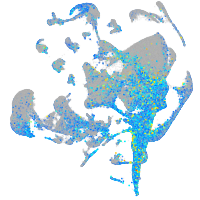
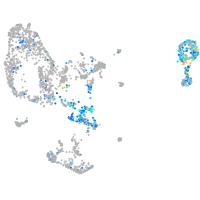
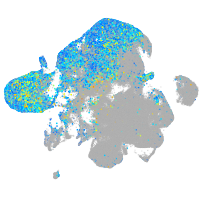
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| zgc:110425 | 0.576 | tob1b | -0.195 |
| mki67 | 0.555 | pvalb1 | -0.170 |
| mibp | 0.554 | tmem59 | -0.162 |
| zgc:165555.3 | 0.552 | COX3 | -0.161 |
| si:dkey-261m9.17 | 0.545 | pvalb2 | -0.160 |
| ccna2 | 0.544 | gapdhs | -0.158 |
| si:ch211-113a14.18 | 0.538 | spint2 | -0.156 |
| cks1b | 0.533 | b2ml | -0.155 |
| mad2l1 | 0.527 | atp2b1a | -0.151 |
| cdca5 | 0.515 | mt-co2 | -0.149 |
| cdk1 | 0.513 | rnasekb | -0.148 |
| aurkb | 0.511 | pvalb8 | -0.144 |
| rrm1 | 0.508 | atp6v1e1b | -0.143 |
| si:dkey-108k21.10 | 0.497 | eef1da | -0.141 |
| dek | 0.495 | calm1a | -0.140 |
| fbxo5 | 0.494 | eno1a | -0.139 |
| ncapg | 0.483 | pkig | -0.136 |
| dut | 0.476 | CR383676.1 | -0.134 |
| rrm2 | 0.467 | actc1b | -0.134 |
| top2a | 0.467 | gpx2 | -0.132 |
| zgc:153405 | 0.463 | si:dkey-16p21.8 | -0.132 |
| smc2 | 0.459 | nupr1a | -0.132 |
| lbr | 0.454 | atp6ap2 | -0.131 |
| hmgb2a | 0.451 | nptna | -0.131 |
| si:dkey-6i22.5 | 0.448 | calm1b | -0.130 |
| tpx2 | 0.446 | wbp2nl | -0.130 |
| stmn1a | 0.445 | socs3a | -0.129 |
| dhfr | 0.445 | gabarapa | -0.129 |
| chaf1a | 0.442 | otofb | -0.129 |
| plk1 | 0.439 | dedd1 | -0.129 |
| hist1h4l | 0.437 | cd164l2 | -0.129 |
| ncapd2 | 0.437 | apoa2 | -0.128 |
| pcna | 0.436 | XLOC-043857 | -0.128 |
| tubb2b | 0.433 | prr15la | -0.128 |
| h2afx | 0.429 | calml4a | -0.128 |