zgc:101130
ZFIN 
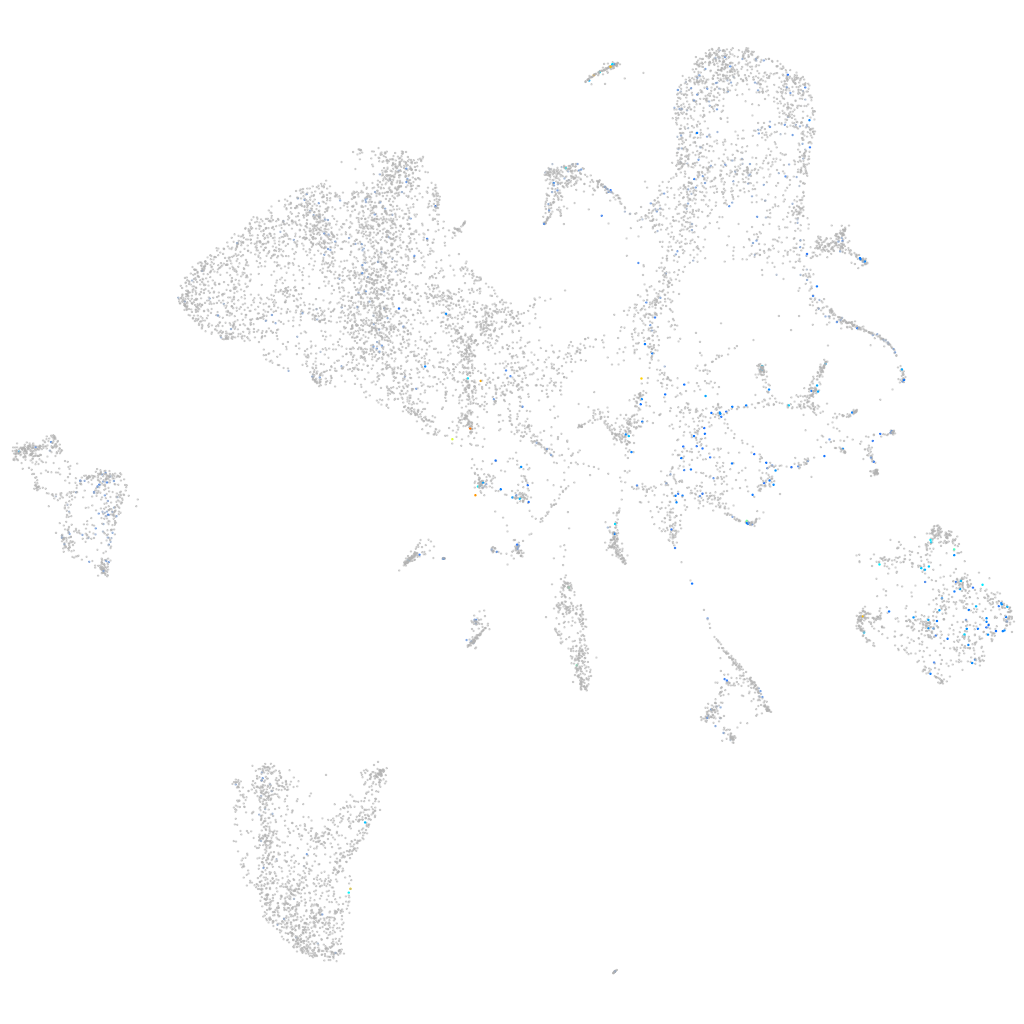
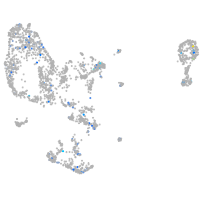
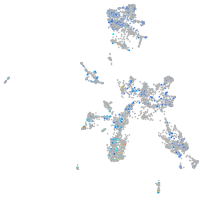
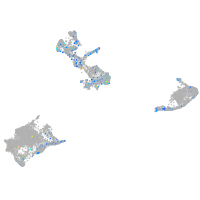
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| CABZ01118767.1 | 0.198 | gapdh | -0.096 |
| col28a2b | 0.188 | ahcy | -0.095 |
| wu:fj39g12 | 0.179 | gatm | -0.095 |
| fut9a | 0.171 | gamt | -0.093 |
| XLOC-022269 | 0.168 | bhmt | -0.086 |
| kcnk15 | 0.168 | fbp1b | -0.084 |
| XLOC-023835 | 0.167 | mat1a | -0.083 |
| nmur3 | 0.163 | agxtb | -0.082 |
| kcnc3b | 0.159 | nupr1b | -0.080 |
| lrrc18b | 0.156 | scp2a | -0.079 |
| syngap1a | 0.155 | aqp12 | -0.077 |
| LOC103910840 | 0.152 | apoa1b | -0.077 |
| FO818722.1 | 0.149 | aldh6a1 | -0.077 |
| si:zfos-364h11.1 | 0.147 | aldob | -0.077 |
| znf1002 | 0.146 | apoa4b.1 | -0.075 |
| ucmaa | 0.145 | fabp10a | -0.074 |
| si:dkeyp-74b6.2 | 0.139 | zgc:92744 | -0.074 |
| diras1b | 0.131 | pgm1 | -0.074 |
| si:ch211-158d24.2 | 0.131 | glud1b | -0.074 |
| atp8b4 | 0.130 | apoa2 | -0.073 |
| neurod2 | 0.129 | dap | -0.073 |
| si:ch211-197n1.2 | 0.128 | grhprb | -0.073 |
| si:ch211-8c17.2 | 0.123 | eno3 | -0.073 |
| BX649423.2 | 0.122 | serpina1 | -0.072 |
| hmgb3a | 0.120 | ttc36 | -0.072 |
| LOC103910768 | 0.119 | agxta | -0.071 |
| gabra1 | 0.119 | fabp3 | -0.071 |
| XLOC-015481 | 0.119 | hpda | -0.071 |
| mir132-1 | 0.117 | rbp2b | -0.071 |
| asic1a | 0.117 | tfa | -0.070 |
| LOC100330186 | 0.116 | pnp4b | -0.070 |
| slc8a4a | 0.116 | nipsnap3a | -0.070 |
| galr2a | 0.116 | ndrg2 | -0.070 |
| marcksl1b | 0.115 | serpina1l | -0.070 |
| hmgn6 | 0.115 | cx32.3 | -0.069 |