zinc finger E-box binding homeobox 2b
ZFIN 
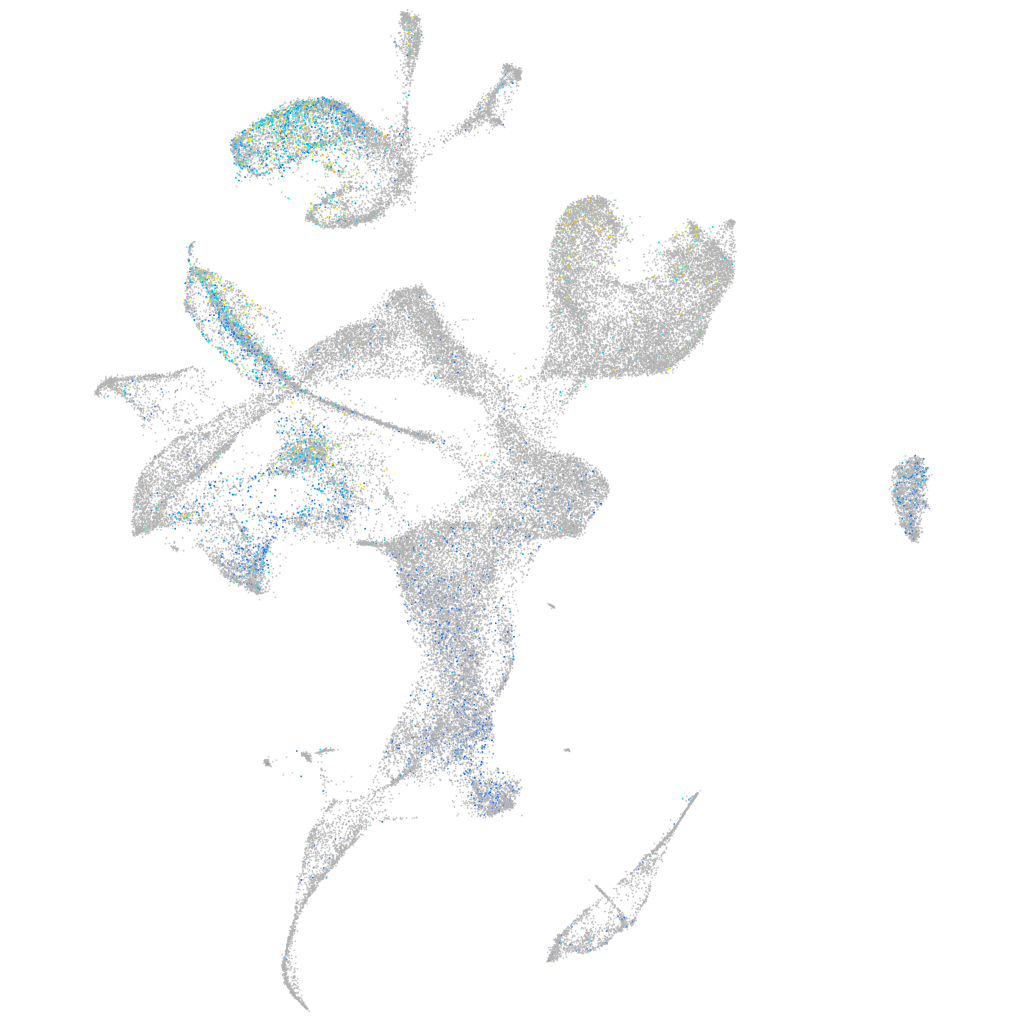
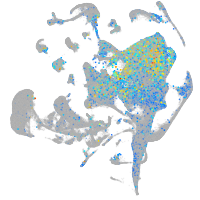
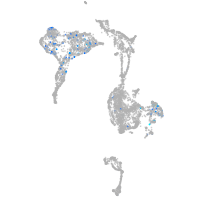
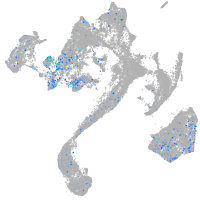
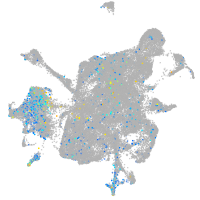
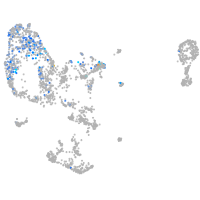
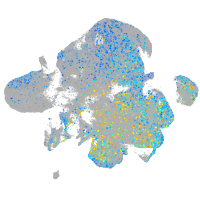
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| stxbp1a | 0.231 | hmgb2a | -0.163 |
| rtn1b | 0.228 | fabp7a | -0.100 |
| elavl3 | 0.227 | msi1 | -0.088 |
| tfap2e | 0.226 | ahcy | -0.088 |
| ppp1r14ba | 0.214 | otx5 | -0.081 |
| stx1b | 0.208 | mdka | -0.078 |
| gng3 | 0.207 | crx | -0.078 |
| ywhag2 | 0.205 | stmn1a | -0.076 |
| zgc:153426 | 0.205 | ddah2 | -0.075 |
| eno2 | 0.204 | ndrg1b | -0.073 |
| pax10 | 0.204 | lbr | -0.073 |
| slc32a1 | 0.202 | neurod4 | -0.072 |
| mir181b-3 | 0.201 | rrm1 | -0.067 |
| myt1la | 0.198 | dut | -0.067 |
| zgc:65894 | 0.196 | msna | -0.067 |
| syt1a | 0.195 | pcna | -0.066 |
| LOC101886114 | 0.195 | hes6 | -0.066 |
| celf5a | 0.194 | tuba8l | -0.065 |
| pbx1a | 0.193 | eef1da | -0.065 |
| tfap2c | 0.191 | dlgap5 | -0.064 |
| sncb | 0.188 | mki67 | -0.064 |
| meis2b | 0.186 | histh1l | -0.064 |
| stmn1b | 0.185 | ccna2 | -0.064 |
| slc35g2b | 0.185 | COX7A2 (1 of many) | -0.062 |
| slc6a1a | 0.185 | nutf2l | -0.061 |
| stmn2a | 0.184 | ccng1 | -0.061 |
| tfap2a | 0.183 | hmga1a | -0.060 |
| olfm2a | 0.179 | sept4a | -0.059 |
| elavl4 | 0.178 | prdm1b | -0.059 |
| sv2a | 0.178 | hmgb2b | -0.059 |
| pbx3b | 0.175 | mad2l1 | -0.058 |
| tfap2b | 0.174 | cdc20 | -0.058 |
| si:ch211-129p13.1 | 0.172 | dek | -0.058 |
| mab21l1 | 0.171 | znf516 | -0.058 |
| c1ql1a | 0.170 | tspan7 | -0.057 |