zinc finger E-box binding homeobox 2a
ZFIN 
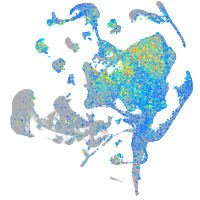
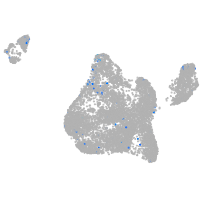
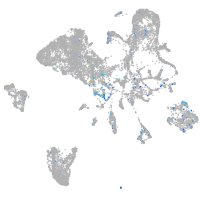
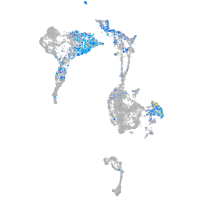
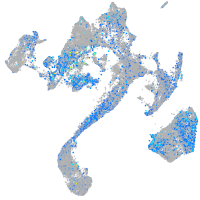
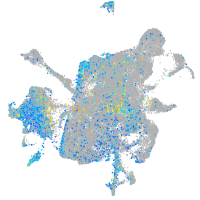
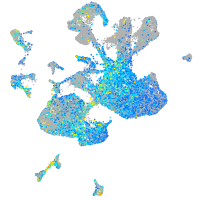
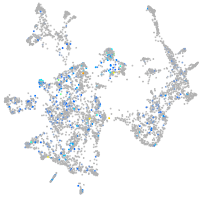
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| nova2 | 0.154 | pkmb | -0.111 |
| tmsb | 0.143 | si:ch211-255p10.3 | -0.109 |
| dla | 0.139 | si:dkey-16p21.8 | -0.105 |
| tuba1a | 0.134 | ckmt2b | -0.100 |
| mdkb | 0.129 | nme2b.2 | -0.098 |
| efnb2b | 0.124 | casq1b | -0.094 |
| hnrnpa1a | 0.122 | adssl1 | -0.092 |
| si:ch211-137a8.4 | 0.120 | ampd1 | -0.092 |
| tardbp | 0.120 | ugp2a | -0.092 |
| notch3 | 0.119 | angptl7 | -0.091 |
| cirbpa | 0.117 | eef2l2 | -0.090 |
| her4.2 | 0.117 | actn3a | -0.088 |
| si:dkey-56m19.5 | 0.115 | slc25a4 | -0.088 |
| ilf3b | 0.114 | smyd1a | -0.088 |
| NC-002333.4 | 0.113 | myoc | -0.087 |
| neurod4 | 0.113 | XLOC-006515 | -0.087 |
| sfrp1a | 0.112 | CABZ01061524.1 | -0.087 |
| acin1a | 0.111 | dhrs7cb | -0.087 |
| ncl | 0.111 | pfkma | -0.086 |
| ppiaa | 0.111 | pvalb1 | -0.086 |
| nop56 | 0.110 | myom1b | -0.085 |
| marcksb | 0.110 | twist1a | -0.084 |
| top1l | 0.110 | XLOC-025819 | -0.084 |
| gpm6aa | 0.108 | XLOC-005350 | -0.084 |
| ppp2cb | 0.107 | igfn1.1 | -0.083 |
| abcf1 | 0.106 | fbp2 | -0.082 |
| dlb | 0.106 | slc25a55b | -0.082 |
| hnrnpub | 0.106 | hsc70 | -0.082 |
| akap12b | 0.105 | col5a2a | -0.081 |
| tuba1c | 0.105 | xirp2b | -0.081 |
| safb | 0.105 | pgam2 | -0.080 |
| hnrnpm | 0.105 | rpl37 | -0.080 |
| rbm4.3 | 0.105 | pvalb4 | -0.080 |
| chd7 | 0.104 | mfap2 | -0.080 |
| hnrnpabb | 0.104 | cdnf | -0.080 |