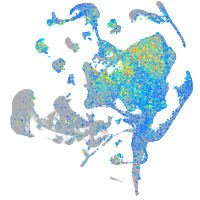
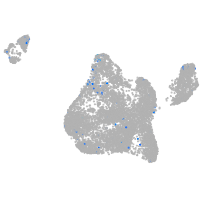
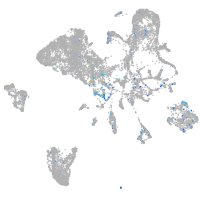
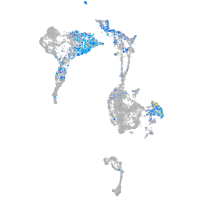
zinc finger E-box binding homeobox 2a
ZFIN 
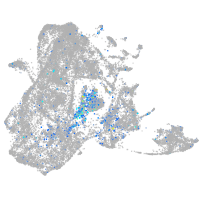
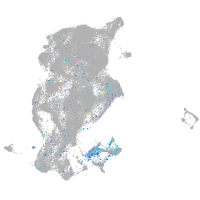
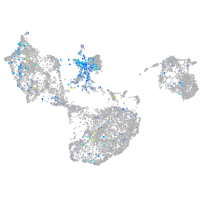
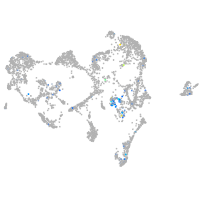
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| mir219-3 | 0.229 | aldob | -0.100 |
| XLOC-003690 | 0.218 | dap | -0.096 |
| efcc1 | 0.217 | gapdh | -0.094 |
| pax3a | 0.215 | aldh6a1 | -0.092 |
| XLOC-003689 | 0.212 | eno3 | -0.090 |
| mdka | 0.204 | cx32.3 | -0.088 |
| LOC798783 | 0.203 | suclg1 | -0.088 |
| si:ch211-57n23.4 | 0.186 | nupr1b | -0.088 |
| gpr17 | 0.179 | mat1a | -0.088 |
| inavaa | 0.179 | gstt1a | -0.087 |
| nova2 | 0.178 | fbp1b | -0.085 |
| BX664610.1 | 0.177 | pnp4b | -0.085 |
| tuba1c | 0.175 | gamt | -0.085 |
| gfap | 0.171 | abat | -0.084 |
| XLOC-003692 | 0.168 | glud1b | -0.084 |
| qkib | 0.167 | scp2a | -0.084 |
| rfx4 | 0.166 | pgk1 | -0.083 |
| gpm6bb | 0.166 | ahcy | -0.083 |
| zgc:165461 | 0.164 | sod1 | -0.082 |
| elavl3 | 0.161 | cox7a1 | -0.082 |
| cyth3b | 0.158 | gstr | -0.081 |
| BX323067.1 | 0.157 | pklr | -0.081 |
| si:ch73-21g5.7 | 0.157 | etnppl | -0.080 |
| cldn5a | 0.156 | nipsnap3a | -0.080 |
| irx1a | 0.154 | gatm | -0.080 |
| ompa | 0.154 | sult2st2 | -0.079 |
| her4.2 | 0.152 | cat | -0.078 |
| si:ch211-218m3.11 | 0.152 | agxtb | -0.078 |
| plp1a | 0.148 | aqp12 | -0.078 |
| abhd6a | 0.148 | g6pca.2 | -0.077 |
| gadd45gb.1 | 0.146 | ckba | -0.077 |
| marcksl1a | 0.146 | slc37a4a | -0.077 |
| XLOC-017216 | 0.146 | upb1 | -0.077 |
| cntnap1 | 0.146 | agxta | -0.076 |
| CR751602.2 | 0.145 | hdlbpa | -0.076 |