zinc finger and BTB domain containing 42
ZFIN 
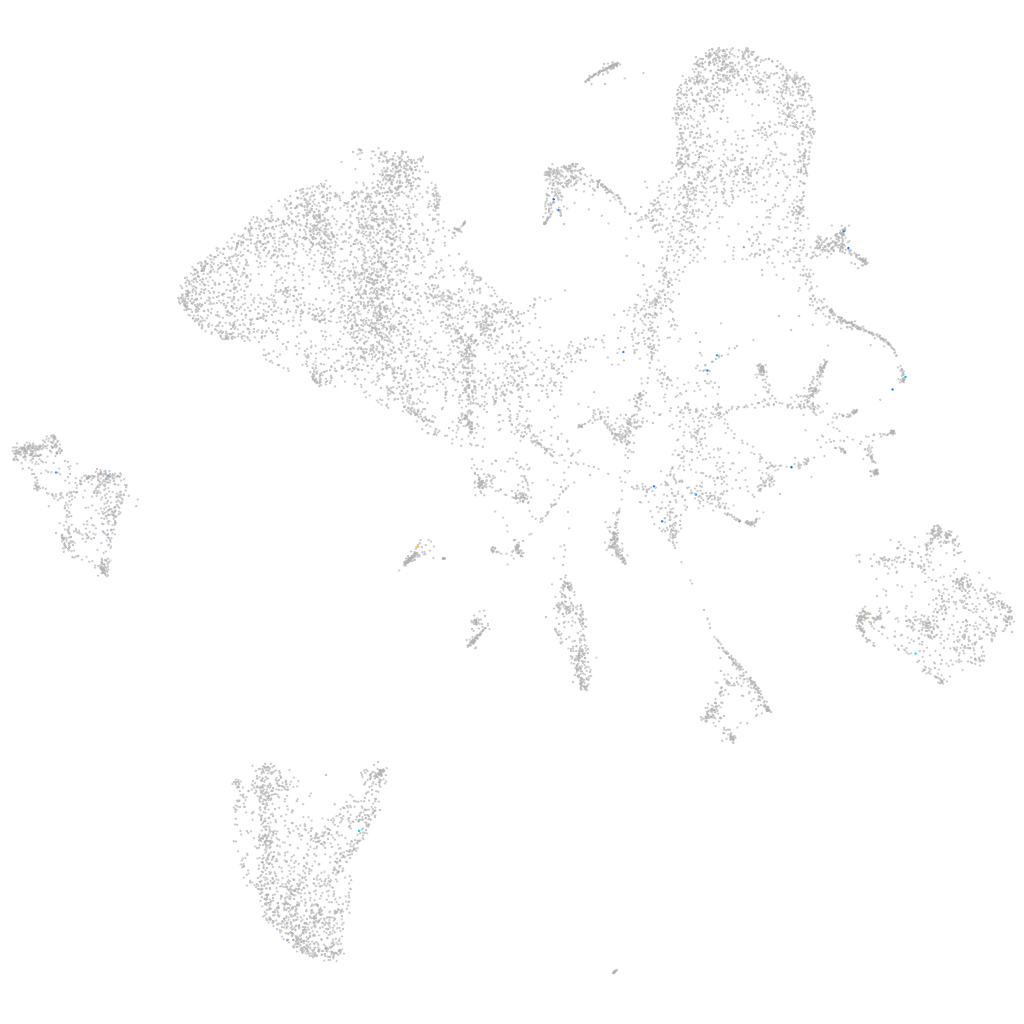
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| glsl | 0.363 | gapdh | -0.044 |
| XLOC-026004 | 0.291 | atp5mc1 | -0.041 |
| tapbpl | 0.270 | eno3 | -0.041 |
| BX936284.1 | 0.220 | gamt | -0.039 |
| BX539313.2 | 0.180 | glud1b | -0.039 |
| CT573446.1 | 0.171 | gpx4a | -0.039 |
| CU672266.1 | 0.153 | atp5f1d | -0.037 |
| CABZ01066611.1 | 0.152 | atp5pf | -0.037 |
| gpr156 | 0.142 | apoa1b | -0.037 |
| stpg2 | 0.137 | cox4i1 | -0.036 |
| si:ch211-93e11.8 | 0.135 | suclg1 | -0.036 |
| lmcd1 | 0.132 | ahcy | -0.036 |
| pimr125 | 0.128 | scp2a | -0.036 |
| BX248511.1 | 0.123 | apoa4b.1 | -0.035 |
| LOC101883960 | 0.122 | apoa2 | -0.035 |
| XLOC-031824 | 0.120 | gatm | -0.035 |
| iffo2b | 0.119 | fbp1b | -0.034 |
| mocs2 | 0.118 | apoc2 | -0.034 |
| ikbke | 0.118 | pgk1 | -0.034 |
| tspearb | 0.118 | bhmt | -0.034 |
| CR450786.1 | 0.117 | afp4 | -0.034 |
| gria1b | 0.117 | gstt1a | -0.034 |
| ntrk2a | 0.114 | cox7a2a | -0.033 |
| CU571091.1 | 0.113 | grhprb | -0.033 |
| LOC110439398 | 0.112 | nipsnap3a | -0.033 |
| CU694808.1 | 0.112 | tfa | -0.032 |
| ANO7 | 0.111 | cyb5a | -0.032 |
| mapta | 0.109 | pklr | -0.032 |
| filip1l | 0.107 | suclg2 | -0.032 |
| sh3bgr | 0.106 | mat1a | -0.032 |
| tcap | 0.106 | acmsd | -0.031 |
| adamtsl2 | 0.104 | prdx2 | -0.031 |
| zgc:153142 | 0.104 | zgc:123103 | -0.031 |
| ap5b1 | 0.103 | apom | -0.031 |
| si:ch73-158p21.2 | 0.102 | rbp2b | -0.031 |