sperm-tail PG-rich repeat containing 2
ZFIN 
Other cell groups


all cells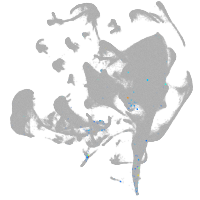 |
blastomeres |
PGCs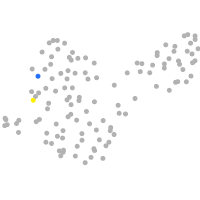 |
endoderm |
axial mesoderm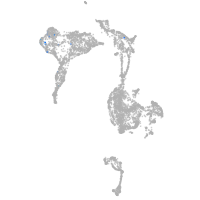 |
muscle  |
mural cells / non-skeletal muscle 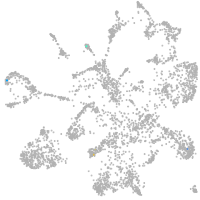 |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| tekt2 | 0.127 | gpm6aa | -0.019 |
| zgc:195356 | 0.117 | gpm6ab | -0.017 |
| gb:eh456644 | 0.106 | rtn1a | -0.017 |
| si:ch211-147k9.8 | 0.097 | hbbe1.3 | -0.016 |
| tekt1 | 0.094 | nova2 | -0.016 |
| tekt4 | 0.094 | stmn1b | -0.016 |
| fam228a | 0.093 | actc1b | -0.015 |
| CR847897.1 | 0.086 | gng3 | -0.015 |
| iqcg | 0.085 | hbae1.1 | -0.015 |
| spata18 | 0.085 | hbae3 | -0.015 |
| CFAP77 | 0.084 | pvalb1 | -0.015 |
| capsla | 0.082 | pvalb2 | -0.015 |
| zgc:153738 | 0.081 | atp6v0cb | -0.014 |
| ak9 | 0.080 | cspg5a | -0.014 |
| rsph14 | 0.079 | mdkb | -0.014 |
| ccdc78 | 0.078 | ndrg4 | -0.014 |
| ppp1r42 | 0.077 | snap25b | -0.014 |
| zgc:153146 | 0.077 | tuba1c | -0.014 |
| armc3 | 0.076 | ywhag2 | -0.014 |
| mycbpap | 0.076 | marcksl1a | -0.014 |
| kif9 | 0.075 | elavl3 | -0.013 |
| ppil6 | 0.075 | elavl4 | -0.013 |
| tbata | 0.075 | hbbe1.1 | -0.013 |
| casc1 | 0.074 | rtn1b | -0.013 |
| ANKRD66 | 0.073 | stx1b | -0.013 |
| dnah5l | 0.073 | sypb | -0.013 |
| LOC108181470 | 0.073 | atp1a3a | -0.012 |
| rsph3 | 0.073 | atp1b2a | -0.012 |
| ak7b | 0.072 | celf2 | -0.012 |
| LOC100329490 | 0.072 | fez1 | -0.012 |
| si:dkey-11c5.11 | 0.072 | gap43 | -0.012 |
| theg | 0.072 | gng2 | -0.012 |
| zgc:153142 | 0.072 | hbbe1.2 | -0.012 |
| cfap206 | 0.070 | sncgb | -0.012 |
| cfap43 | 0.070 | stmn2a | -0.012 |




