peptidylprolyl isomerase (cyclophilin)-like 6
ZFIN 
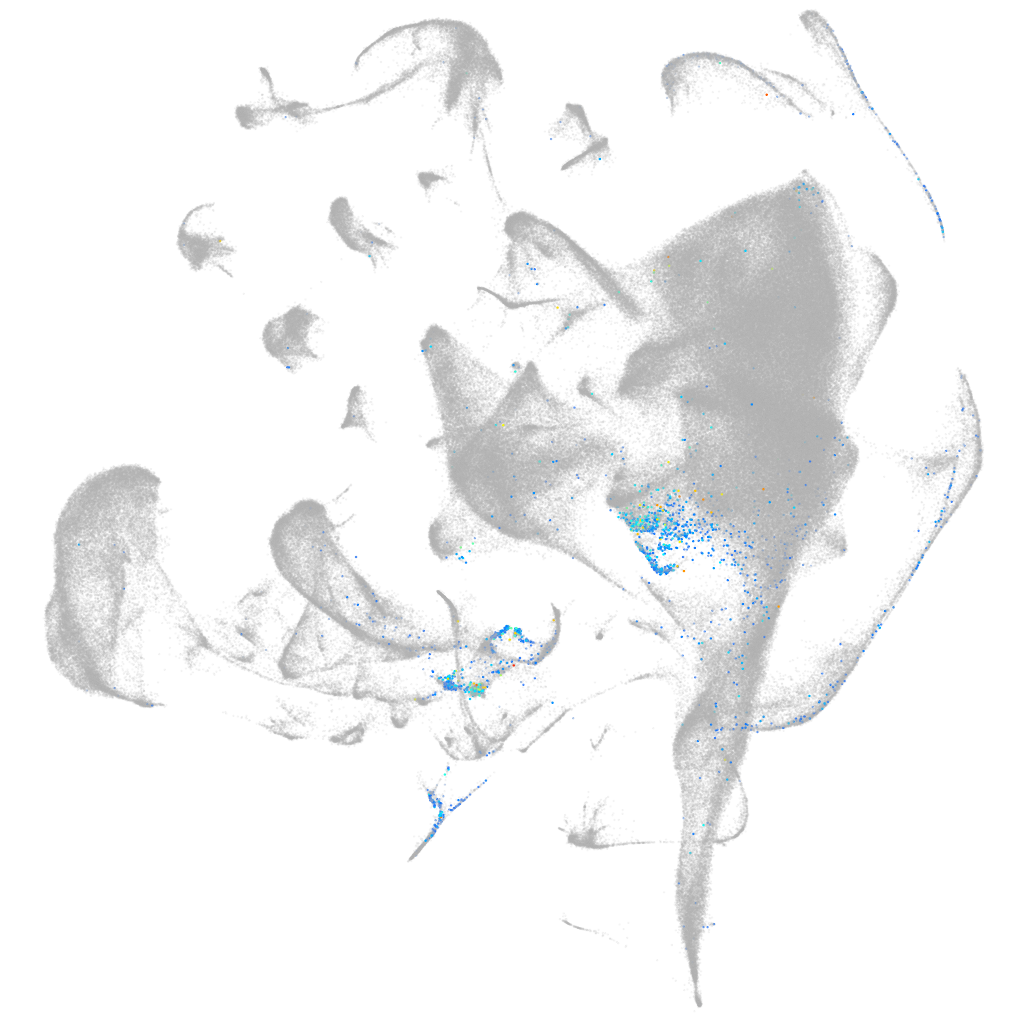
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm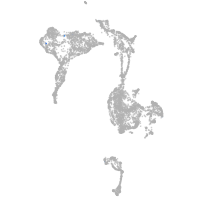 |
muscle 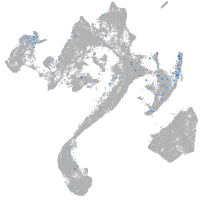 |
mural cells / non-skeletal muscle  |
mesenchyme 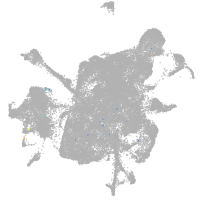 |
hematopoietic / vasculature  |
pronephros 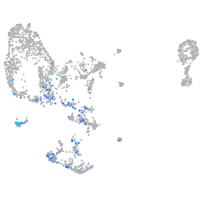 |
neural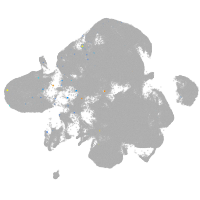 |
spinal cord / glia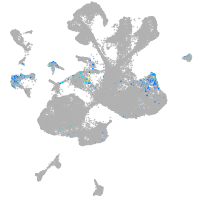 |
eye |
taste / olfactory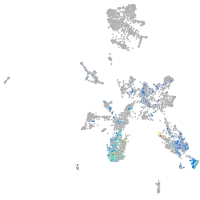 |
otic / lateral line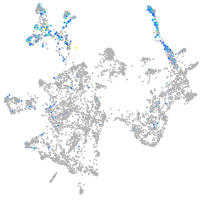 |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| tekt1 | 0.276 | gpm6ab | -0.032 |
| enkur | 0.269 | celf2 | -0.031 |
| pacrg | 0.256 | gpm6aa | -0.027 |
| tekt4 | 0.254 | tubb5 | -0.025 |
| capsla | 0.253 | rbfox1 | -0.023 |
| smkr1 | 0.250 | rtn1b | -0.023 |
| iqcg | 0.248 | stmn1b | -0.023 |
| CFAP77 | 0.246 | cadm3 | -0.022 |
| spag6 | 0.246 | tmem59l | -0.022 |
| tbata | 0.246 | elavl4 | -0.021 |
| ropn1l | 0.245 | fez1 | -0.021 |
| si:dkey-148f10.4 | 0.245 | gap43 | -0.021 |
| daw1 | 0.242 | mab21l1 | -0.021 |
| fam228a | 0.241 | myt1b | -0.021 |
| tekt2 | 0.241 | ndrg4 | -0.021 |
| ccdc151 | 0.239 | nova1 | -0.021 |
| si:dkey-27p23.3 | 0.237 | scrt2 | -0.021 |
| cfap126 | 0.235 | snap25b | -0.021 |
| spata18 | 0.235 | ywhag2 | -0.021 |
| dnali1 | 0.231 | zgc:153426 | -0.021 |
| mycbpap | 0.229 | cyt1l | -0.020 |
| lrrc23 | 0.227 | hbbe1.3 | -0.020 |
| capslb | 0.223 | krtt1c19e | -0.020 |
| ccdc114 | 0.222 | nova2 | -0.020 |
| cfap52 | 0.221 | sncgb | -0.020 |
| nme5 | 0.221 | tmsb2 | -0.020 |
| si:ch211-163l21.7 | 0.221 | gabrb4 | -0.020 |
| rsph14 | 0.216 | atp1b2a | -0.019 |
| foxj1a | 0.214 | camk2d2 | -0.019 |
| morn3 | 0.214 | cyt1 | -0.019 |
| got1l1 | 0.213 | elavl3 | -0.019 |
| ak7a | 0.212 | elmo1 | -0.019 |
| ppp1r42 | 0.212 | hbae3 | -0.019 |
| gb:eh456644 | 0.210 | mab21l2 | -0.019 |
| spaca9 | 0.208 | si:ch73-386h18.1 | -0.019 |