"3-monooxygenase/tryptophan 5-monooxygenase activation protein, gamma polypeptide 1"
ZFIN 
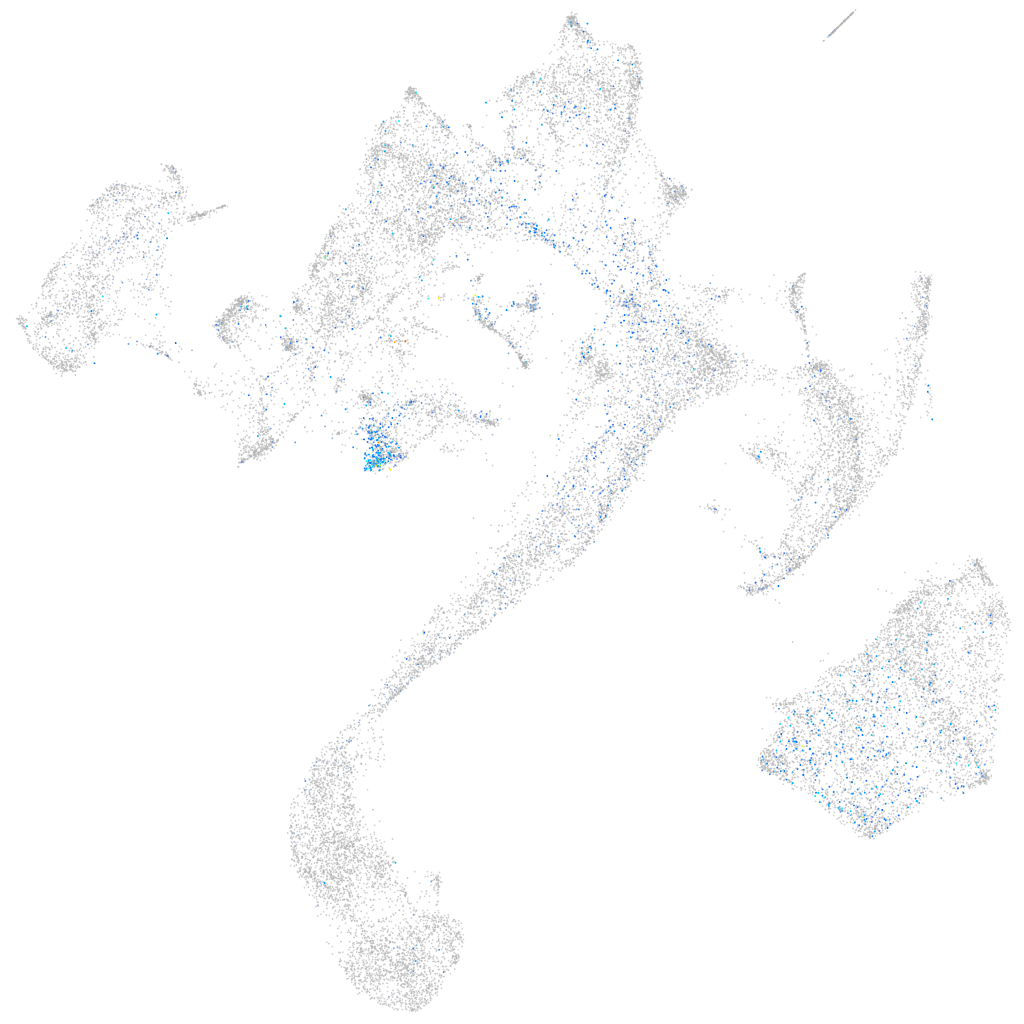
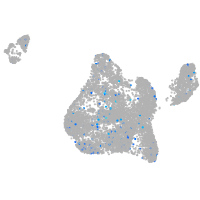
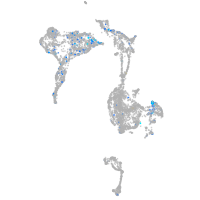
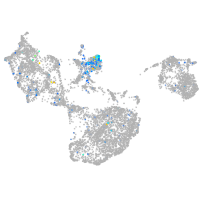
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| sncb | 0.216 | aldoab | -0.082 |
| snap25a | 0.214 | acta1b | -0.078 |
| vamp2 | 0.213 | actc1b | -0.078 |
| cdk5r2a | 0.212 | tmem38a | -0.077 |
| stxbp1a | 0.211 | mybphb | -0.077 |
| zgc:65894 | 0.210 | tpma | -0.077 |
| rtn1b | 0.209 | tnnc2 | -0.077 |
| stx1b | 0.205 | ttn.1 | -0.076 |
| gng3 | 0.202 | ckmb | -0.075 |
| eno2 | 0.202 | ckma | -0.075 |
| stmn2a | 0.202 | atp5mc3b | -0.073 |
| si:dkeyp-72g9.4 | 0.199 | hhatla | -0.073 |
| zgc:153426 | 0.198 | actn3b | -0.072 |
| gap43 | 0.194 | actn3a | -0.072 |
| stmn1b | 0.194 | ldb3a | -0.072 |
| elavl4 | 0.193 | atp2a1 | -0.071 |
| olfm1b | 0.187 | tmod4 | -0.071 |
| cspg5a | 0.182 | ldb3b | -0.070 |
| gpm6aa | 0.182 | neb | -0.070 |
| myt1la | 0.180 | actc1a | -0.070 |
| basp1 | 0.178 | ttn.2 | -0.069 |
| tuba2 | 0.175 | si:ch211-255p10.3 | -0.068 |
| map1aa | 0.174 | myom1a | -0.068 |
| vsnl1b | 0.171 | smyd1a | -0.068 |
| sv2a | 0.170 | tnnt3a | -0.068 |
| sypa | 0.169 | srl | -0.068 |
| cplx2 | 0.168 | ak1 | -0.068 |
| cplx2l | 0.168 | mylpfa | -0.068 |
| evlb | 0.168 | XLOC-025819 | -0.067 |
| gabrg2 | 0.167 | myl1 | -0.067 |
| tuba1c | 0.166 | hsp90aa1.1 | -0.067 |
| atp6v0cb | 0.166 | si:dkey-151g10.6 | -0.067 |
| maptb | 0.166 | desma | -0.067 |
| elmod1 | 0.164 | si:ch73-367p23.2 | -0.067 |
| sncgb | 0.164 | acta1a | -0.067 |