YTH N(6)-methyladenosine RNA binding protein 2
ZFIN 
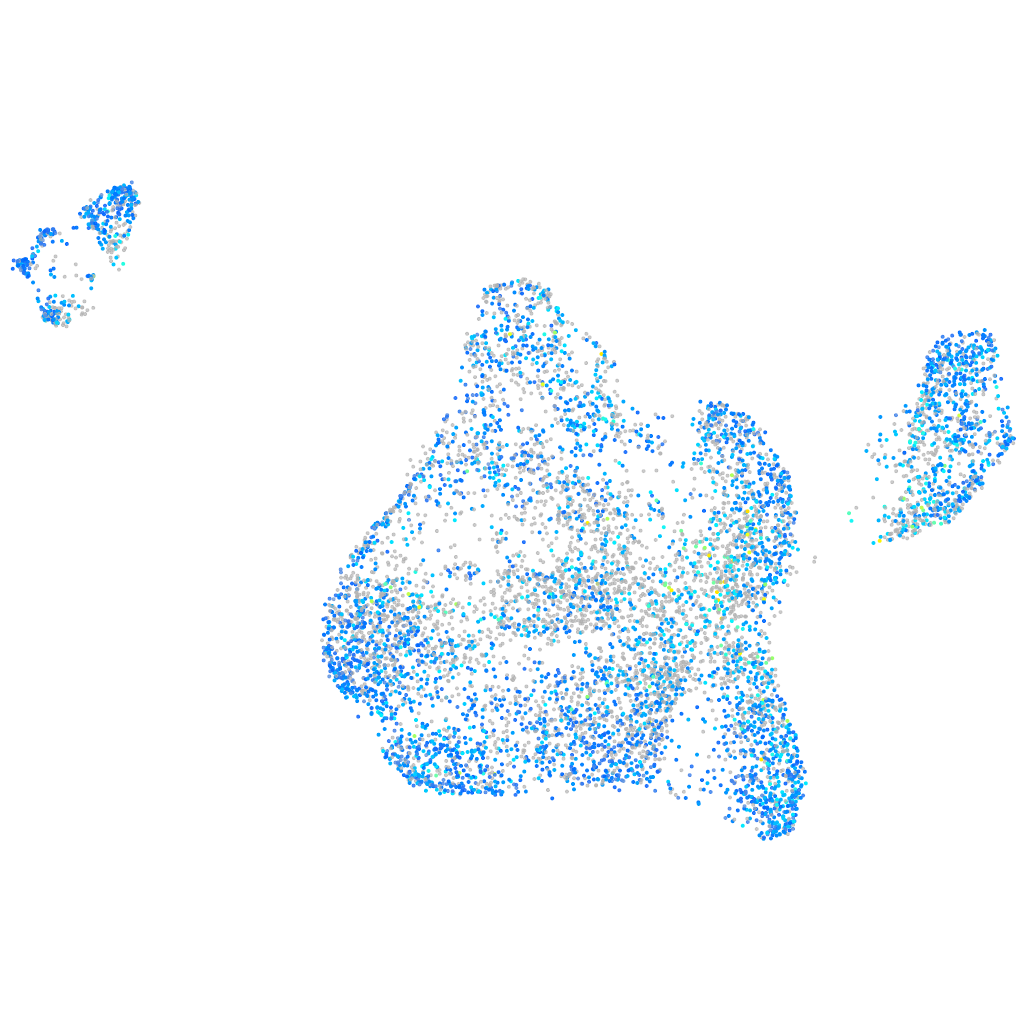
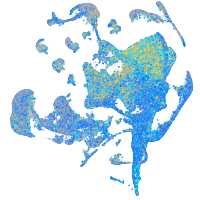
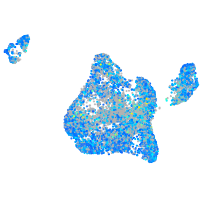
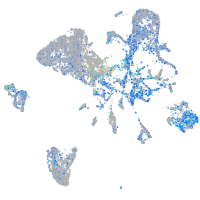
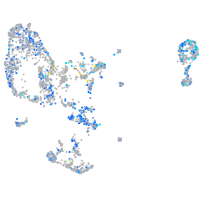
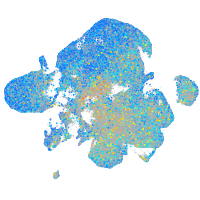
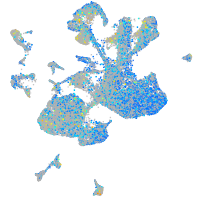
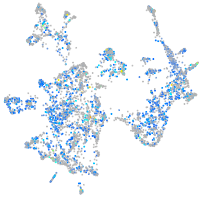
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| mkrn4 | 0.141 | NC-002333.4 | -0.112 |
| gtf2a1 | 0.141 | ptmab | -0.103 |
| dnajc1 | 0.132 | hnrnpa0b | -0.093 |
| nanog | 0.132 | eef1a1l1 | -0.092 |
| h1m | 0.131 | hspb1 | -0.087 |
| rbm7 | 0.129 | rpl7a | -0.082 |
| zgc:100951 | 0.123 | hmgn2 | -0.077 |
| slc16a3 | 0.120 | rplp1 | -0.076 |
| npc2 | 0.119 | rpl7 | -0.073 |
| lamp2 | 0.118 | mt-co2 | -0.072 |
| cldnd | 0.118 | si:ch211-222l21.1 | -0.071 |
| zc3h13 | 0.116 | hmgb2a | -0.071 |
| acp5a | 0.116 | rps6 | -0.069 |
| ccna1 | 0.116 | sox2 | -0.067 |
| gdf3 | 0.113 | ybx1 | -0.067 |
| syncripl | 0.110 | apoc1 | -0.067 |
| rbpms2b | 0.110 | faua | -0.066 |
| inka1a | 0.109 | dynll1 | -0.066 |
| h2af1al | 0.109 | hmgn6 | -0.066 |
| cnot8 | 0.108 | si:ch211-152c2.3 | -0.064 |
| ehmt1b | 0.108 | rps12 | -0.062 |
| prkrip1 | 0.108 | sp5l | -0.062 |
| rcor1 | 0.108 | cdx4 | -0.062 |
| ube2g1a | 0.108 | mt-co1 | -0.062 |
| spink2.1 | 0.107 | rpl27 | -0.060 |
| sox11b | 0.107 | rplp0 | -0.060 |
| pfn2 | 0.107 | ptgdsb.1 | -0.060 |
| fdx1 | 0.107 | add3b | -0.060 |
| zfand5b | 0.106 | rps2 | -0.060 |
| zgc:173742 | 0.105 | mt-cyb | -0.059 |
| ttf1 | 0.105 | rps4x | -0.058 |
| mycla | 0.104 | rpl30 | -0.058 |
| map1lc3c | 0.104 | rps15a | -0.058 |
| reep2 | 0.104 | hmga1a | -0.057 |
| tent5c | 0.104 | rpl15 | -0.057 |