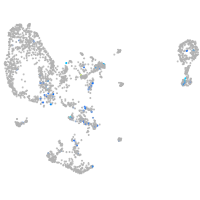
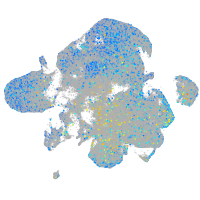
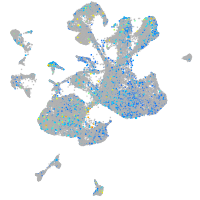
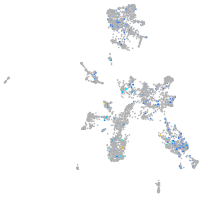
xylosyltransferase I
ZFIN 
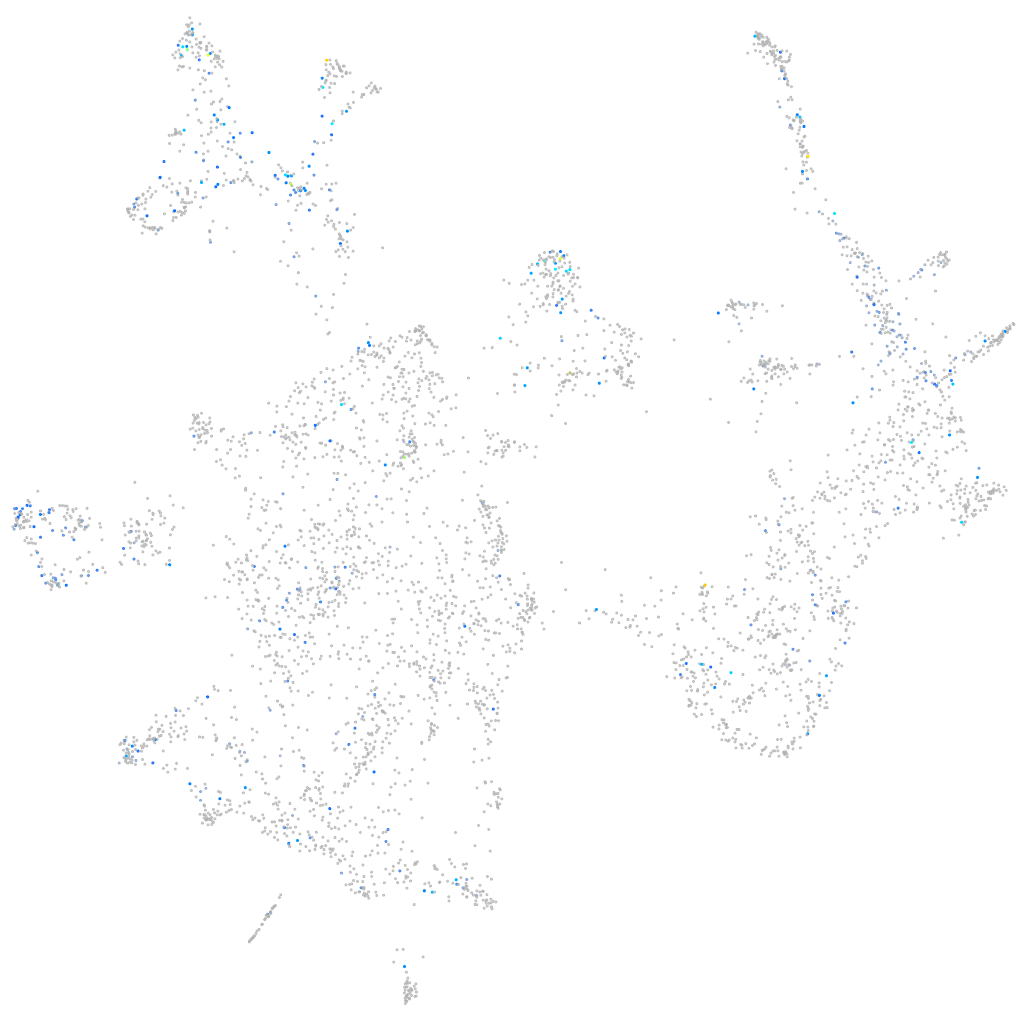
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| zgc:162989 | 0.165 | rpl34 | -0.111 |
| tuba1a | 0.162 | rplp1 | -0.108 |
| si:dkey-280e21.3 | 0.161 | sparc | -0.103 |
| FOXN2 (1 of many) | 0.160 | rpl24 | -0.091 |
| ugt5e1 | 0.156 | rps20 | -0.090 |
| LOC108179813 | 0.154 | epcam | -0.090 |
| CR847844.2 | 0.153 | rpl26 | -0.089 |
| LOC110440222 | 0.152 | si:dkey-151g10.6 | -0.089 |
| nkpd1 | 0.150 | rpl29 | -0.089 |
| baiap2l2b | 0.150 | otomp | -0.088 |
| cacna1da | 0.148 | rps10 | -0.088 |
| sncb | 0.144 | rpl37 | -0.086 |
| si:ch211-250e5.16 | 0.142 | rpl23 | -0.085 |
| CR812470.2 | 0.142 | stm | -0.084 |
| si:dkeyp-80c12.5 | 0.142 | rpl27 | -0.084 |
| meig1 | 0.141 | rps28 | -0.083 |
| otofb | 0.141 | rpl36a | -0.083 |
| myo6b | 0.140 | rpl8 | -0.082 |
| lrrc73 | 0.140 | rpl9 | -0.082 |
| zgc:55461 | 0.140 | rpl14 | -0.082 |
| smpx | 0.140 | rps19 | -0.081 |
| grxcr2 | 0.139 | rpl35a | -0.079 |
| eno1a | 0.138 | rps24 | -0.079 |
| CU861459.1 | 0.137 | rpl38 | -0.079 |
| lhfpl5a | 0.137 | rpl28 | -0.078 |
| mtnr1bb | 0.136 | rpl35 | -0.077 |
| si:ch211-57h10.1 | 0.135 | rplp2l | -0.077 |
| kif1aa | 0.135 | rps26l | -0.076 |
| LOC795227 | 0.135 | rps11 | -0.074 |
| tmc2a | 0.134 | rpl13 | -0.074 |
| atp1a3b | 0.134 | rps27.1 | -0.073 |
| gfi1ab | 0.134 | rps13 | -0.073 |
| vamp2 | 0.134 | rps15a | -0.073 |
| atp2b1a | 0.134 | rps8a | -0.073 |
| nptna | 0.133 | si:ch211-80h18.1 | -0.072 |