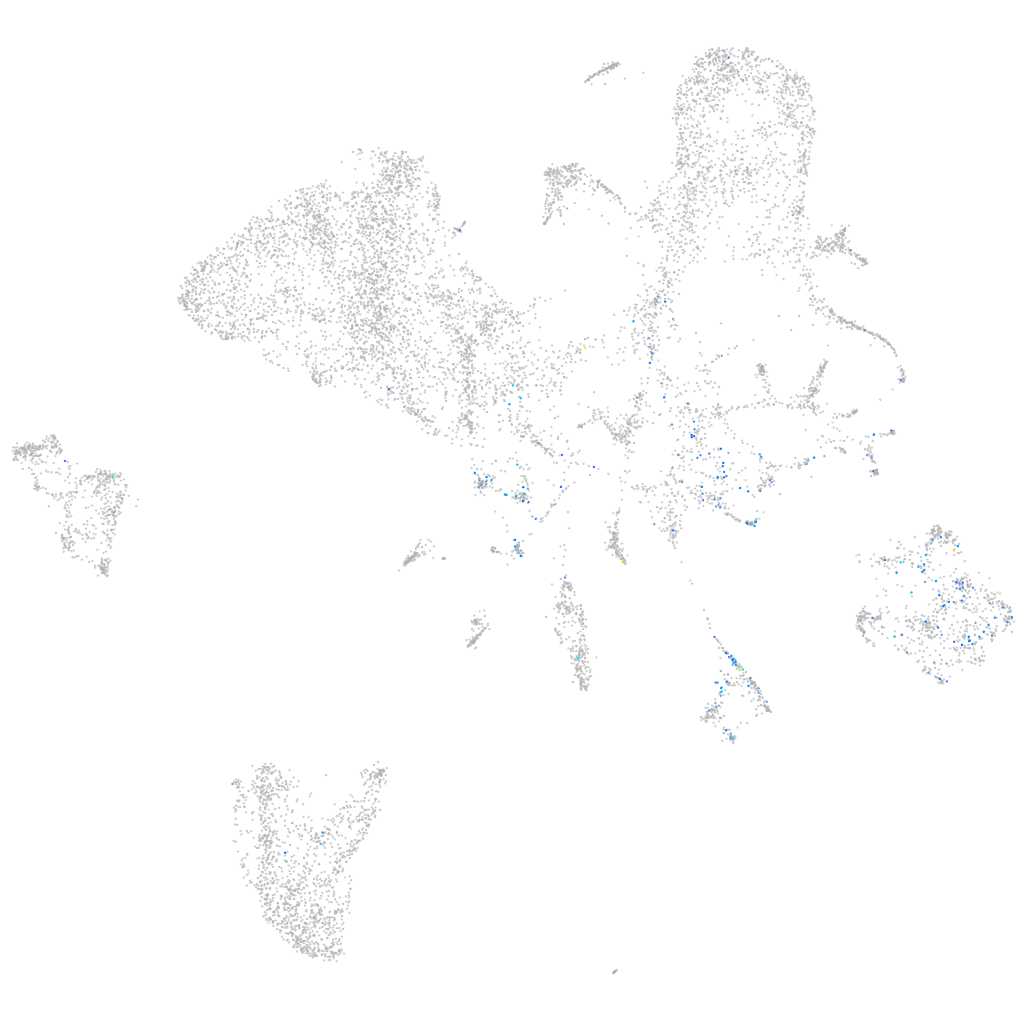
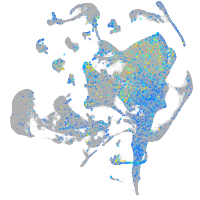
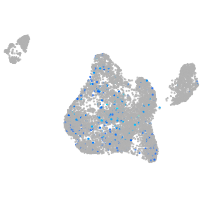
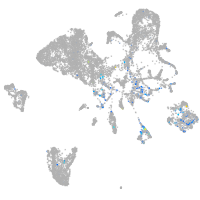
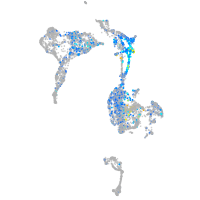
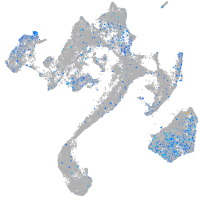
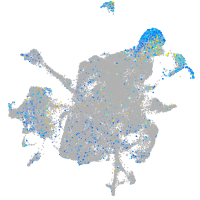
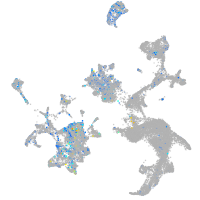
xylosyltransferase I
ZFIN 
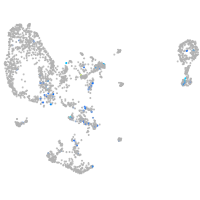
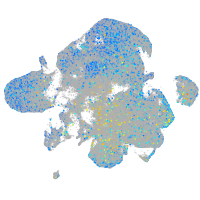
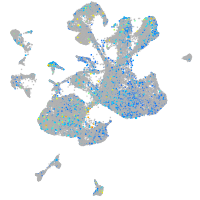
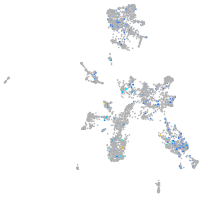
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| lamb1a | 0.176 | gapdh | -0.128 |
| stm | 0.172 | ahcy | -0.126 |
| msna | 0.170 | gamt | -0.121 |
| si:dkey-85n7.8 | 0.165 | aldob | -0.118 |
| cx43.4 | 0.157 | eno3 | -0.114 |
| bcam | 0.157 | dap | -0.113 |
| lamc1 | 0.155 | sod1 | -0.112 |
| sim1b | 0.155 | fbp1b | -0.111 |
| marcksb | 0.150 | gpx4a | -0.109 |
| lama5 | 0.150 | gatm | -0.109 |
| si:ch211-222l21.1 | 0.148 | suclg1 | -0.108 |
| LOC101885599 | 0.147 | atp5if1b | -0.106 |
| ctnnb1 | 0.147 | mat1a | -0.106 |
| khdrbs1a | 0.146 | scp2a | -0.103 |
| akap12b | 0.144 | pklr | -0.101 |
| jpt1b | 0.143 | tpi1b | -0.100 |
| marcksl1a | 0.141 | glud1b | -0.100 |
| marcksl1b | 0.141 | apoa4b.1 | -0.098 |
| tpm4a | 0.141 | mdh1aa | -0.097 |
| XLOC-015433 | 0.140 | pgk1 | -0.095 |
| hmgb3a | 0.139 | apoc2 | -0.095 |
| qkia | 0.138 | abat | -0.095 |
| fstl1b | 0.136 | aldh7a1 | -0.095 |
| ephx4 | 0.135 | gstt1a | -0.095 |
| ppm1g | 0.134 | apoa1b | -0.093 |
| lbr | 0.134 | hadh | -0.093 |
| hmgb1b | 0.134 | aldh6a1 | -0.092 |
| syncrip | 0.134 | tdo2a | -0.092 |
| ntn2 | 0.133 | bhmt | -0.092 |
| cirbpb | 0.133 | rdh1 | -0.091 |
| mnx1 | 0.133 | pnp4b | -0.091 |
| mki67 | 0.133 | prdx2 | -0.091 |
| tgif1 | 0.133 | cx32.3 | -0.091 |
| si:ch73-1a9.3 | 0.133 | suclg2 | -0.091 |
| hmga1a | 0.131 | ugt1a7 | -0.090 |