5'-3' exoribonuclease 2
ZFIN 
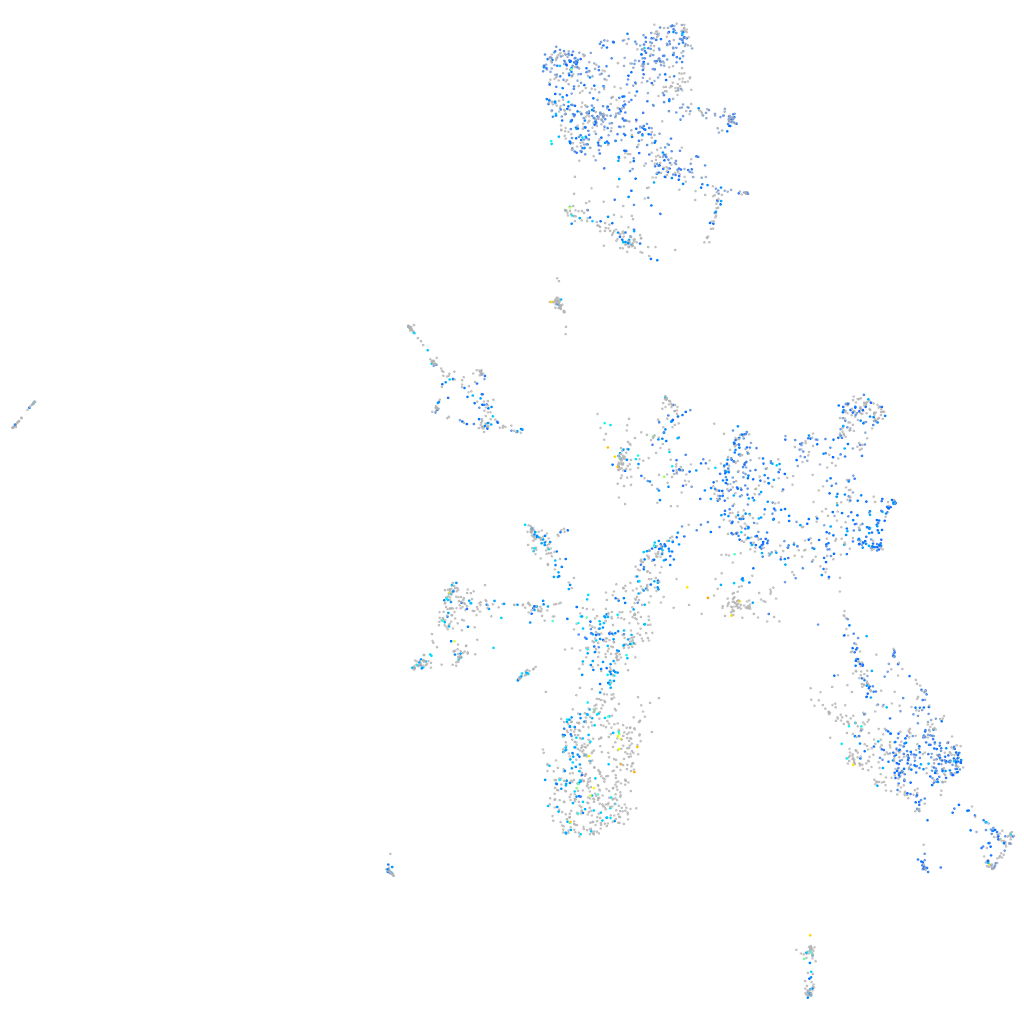
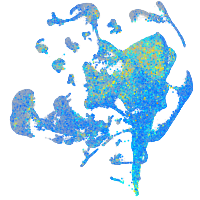
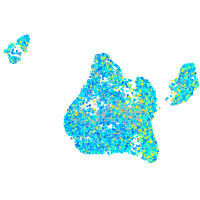
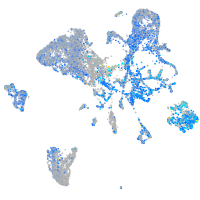
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| seta | 0.158 | pvalb2 | -0.085 |
| setb | 0.154 | hbbe2 | -0.080 |
| hnrnpaba | 0.154 | pvalb1 | -0.075 |
| srsf9 | 0.149 | actc1b | -0.073 |
| khdrbs1a | 0.145 | krtt1c19e | -0.072 |
| cbx3a | 0.144 | apoa1b | -0.072 |
| syncrip | 0.143 | pvalb4 | -0.071 |
| hnrnpabb | 0.140 | ela3l | -0.070 |
| si:ch211-288g17.3 | 0.140 | lgals2b | -0.069 |
| taf13 | 0.138 | hbae3 | -0.063 |
| anp32b | 0.137 | prss1 | -0.059 |
| cct5 | 0.134 | rplp2 | -0.059 |
| smarcd1 | 0.134 | sycn.2 | -0.059 |
| snrpd1 | 0.133 | mtbl | -0.058 |
| sae1 | 0.133 | apoa2 | -0.057 |
| hdac1 | 0.133 | rbp4 | -0.056 |
| hmga1a | 0.133 | col1a1a | -0.056 |
| hmgb2b | 0.132 | icn | -0.055 |
| bud31 | 0.131 | CELA1 (1 of many) | -0.055 |
| sumo3a | 0.129 | mylz3 | -0.054 |
| srsf3b | 0.129 | tnnt3b | -0.053 |
| h2afvb | 0.129 | EARS2 | -0.053 |
| srsf2a | 0.129 | krt5 | -0.053 |
| h3f3a | 0.129 | afp4 | -0.053 |
| snrpf | 0.129 | tfa | -0.053 |
| snrpe | 0.129 | wu:fb18f06 | -0.052 |
| alyref | 0.128 | opn1sw1 | -0.052 |
| tubb2b | 0.127 | dusp26 | -0.051 |
| ddx39ab | 0.125 | ier2b | -0.051 |
| mcm7 | 0.125 | VIT | -0.050 |
| cops2 | 0.125 | gngt2b | -0.050 |
| ran | 0.124 | prss59.1 | -0.050 |
| hnrnpa0l | 0.124 | oat | -0.050 |
| lsm7 | 0.124 | zgc:136930 | -0.050 |
| rbm8a | 0.124 | zgc:112160 | -0.049 |