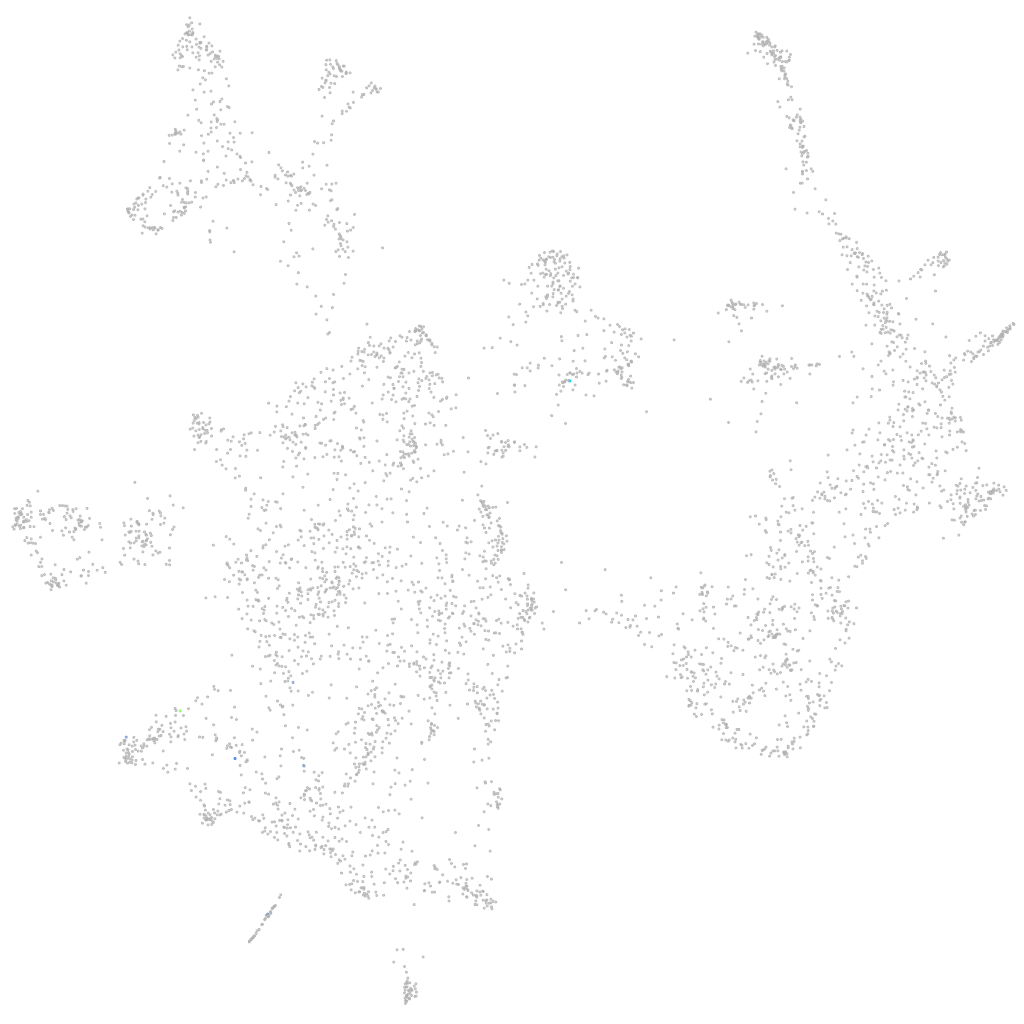
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| XLOC-017492 | 0.643 | h2afx1 | -0.038 |
| XLOC-033456 | 0.586 | cox4i1 | -0.036 |
| CU633486.1 | 0.481 | arf2b | -0.030 |
| arhgap23b | 0.452 | ctsla | -0.030 |
| LOC101886363 | 0.377 | h3f3a | -0.030 |
| BX957326.1 | 0.318 | ier2b | -0.029 |
| pgpep1l | 0.309 | dynll1 | -0.029 |
| tbx5a | 0.307 | calm2a | -0.028 |
| LOC108179174 | 0.302 | mcl1a | -0.028 |
| CU657976.1 | 0.292 | ndufc2 | -0.028 |
| si:dkey-283b15.2 | 0.282 | psmb7 | -0.028 |
| gmip | 0.282 | calm2b | -0.027 |
| galca | 0.272 | tuba8l4 | -0.027 |
| tmem237a | 0.256 | tubb2b | -0.027 |
| diras1b | 0.249 | hmgn7 | -0.027 |
| mtnr1ab | 0.241 | sdc4 | -0.026 |
| arl13a | 0.236 | atp5l | -0.026 |
| echdc1 | 0.225 | atp5mc3b | -0.026 |
| si:dkey-261m9.18 | 0.215 | si:ch1073-325m22.2 | -0.026 |
| mpp4b | 0.214 | dynlrb1 | -0.026 |
| limd1a | 0.209 | cct2 | -0.026 |
| pcdh1g31 | 0.205 | setb | -0.025 |
| slit1a | 0.204 | mpc1 | -0.025 |
| dpagt1 | 0.202 | f11r.1 | -0.025 |
| LOC103909376 | 0.197 | arpc3 | -0.025 |
| si:dkey-16i5.8 | 0.197 | arpc2 | -0.025 |
| cngb3.2 | 0.197 | sec61b | -0.024 |
| zgc:112294 | 0.194 | fkbp1aa | -0.024 |
| ckmt2a | 0.190 | cycsb | -0.024 |
| pcdh8 | 0.189 | cd9b | -0.024 |
| tulp1a | 0.181 | atp5if1a | -0.024 |
| BX119915.2 | 0.177 | cox8a | -0.024 |
| rcvrna | 0.177 | ndufab1b | -0.024 |
| trpv4 | 0.176 | dbi | -0.024 |
| si:ch73-287m6.1 | 0.175 | her15.1 | -0.024 |