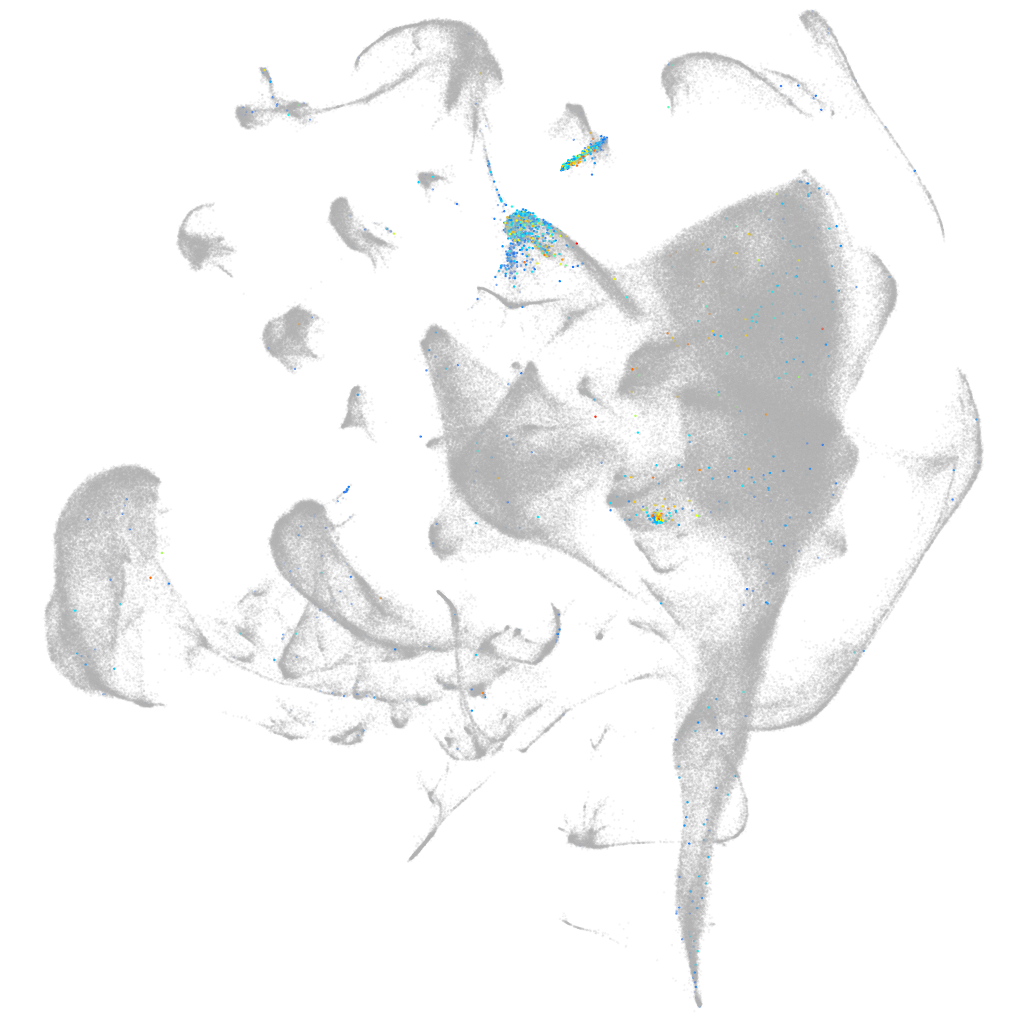Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| msnb | 0.388 | marcksb | -0.041 |
| cracr2ab | 0.343 | rtn1a | -0.041 |
| pmelb | 0.340 | nova2 | -0.037 |
| selenou1b | 0.329 | gpm6aa | -0.036 |
| fgfbp1a | 0.309 | hmgb3a | -0.036 |
| rdh5 | 0.305 | elavl3 | -0.034 |
| rlbp1b | 0.301 | tmeff1b | -0.034 |
| fabp11b | 0.293 | tubb5 | -0.034 |
| rbp4l | 0.293 | zc4h2 | -0.034 |
| pip4k2cb | 0.290 | rtn1b | -0.033 |
| rpe65a | 0.287 | si:ch211-288g17.3 | -0.033 |
| tm6sf2 | 0.287 | stmn1b | -0.033 |
| inpp5kb | 0.282 | marcksl1a | -0.033 |
| slco1e1 | 0.282 | nucks1a | -0.031 |
| best1 | 0.279 | cd99l2 | -0.030 |
| cidea | 0.272 | gng3 | -0.030 |
| abca4a | 0.272 | elavl4 | -0.029 |
| rbp3 | 0.271 | si:dkey-276j7.1 | -0.029 |
| dct | 0.265 | stx1b | -0.029 |
| oca2 | 0.262 | mdkb | -0.028 |
| plin1 | 0.255 | nsg2 | -0.028 |
| pmela | 0.251 | ywhag2 | -0.028 |
| tyrp1b | 0.251 | fscn1a | -0.027 |
| slc4a5b | 0.249 | nova1 | -0.027 |
| pdcb | 0.246 | setb | -0.027 |
| guk1b | 0.242 | syt2a | -0.027 |
| tulp1b | 0.241 | tmsb | -0.027 |
| slc25a3a | 0.240 | stxbp1a | -0.027 |
| rdh8a | 0.239 | hmgb1b | -0.026 |
| rgrb | 0.237 | map1aa | -0.026 |
| rgs9a | 0.237 | nrxn1a | -0.026 |
| si:dkey-17e16.15 | 0.233 | rbfox2 | -0.026 |
| tyrp1a | 0.233 | snap25a | -0.026 |
| gnat2 | 0.232 | sox11b | -0.026 |
| lratb.2 | 0.232 | stmn2a | -0.026 |