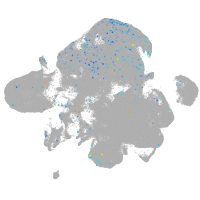
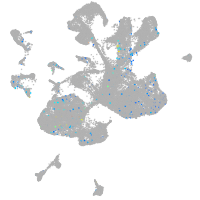
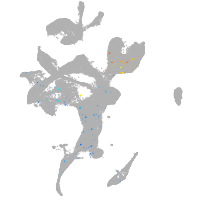
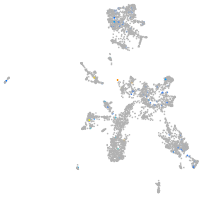
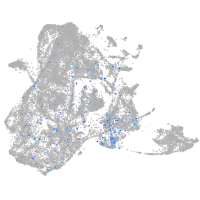
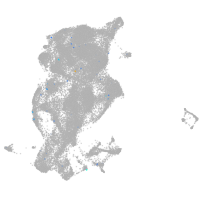
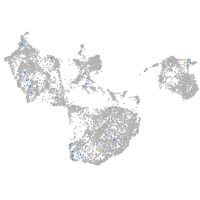
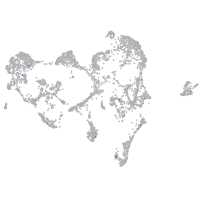
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| CABZ01092072.1 | 0.100 | snap25a | -0.037 |
| pimr203 | 0.072 | gng3 | -0.035 |
| LOC103911458 | 0.072 | elavl4 | -0.035 |
| CT573133.1 | 0.066 | stmn2a | -0.033 |
| zgc:172133 | 0.066 | calm1a | -0.032 |
| NC-002333.21 | 0.066 | ptmaa | -0.032 |
| pimr205 | 0.064 | rtn1b | -0.032 |
| CR855257.2 | 0.063 | sncb | -0.031 |
| LOC110439979 | 0.062 | stmn4l | -0.030 |
| zgc:171452 | 0.059 | gap43 | -0.029 |
| si:ch211-284e13.9 | 0.053 | gabrb4 | -0.028 |
| zgc:171500 | 0.053 | eno2 | -0.028 |
| BX914205.2 | 0.052 | atp1a3a | -0.028 |
| LOC103909765 | 0.051 | ywhag2 | -0.028 |
| AL929576.1 | 0.050 | sncgb | -0.028 |
| CABZ01085878.1 | 0.050 | gpr85 | -0.028 |
| BX119926.1 | 0.048 | syt2a | -0.027 |
| CABZ01066727.1 | 0.046 | evlb | -0.027 |
| LOC101885677 | 0.046 | gjd1a | -0.026 |
| slc1a2a | 0.046 | elavl3 | -0.026 |
| si:ch211-155o21.4 | 0.045 | stx1b | -0.026 |
| wls | 0.044 | csdc2a | -0.025 |
| crtap | 0.044 | zc4h2 | -0.025 |
| sema3gb | 0.043 | rbfox1 | -0.025 |
| sox2 | 0.042 | vamp2 | -0.025 |
| LOC110438454 | 0.042 | zgc:101840 | -0.025 |
| nog2 | 0.041 | isl2a | -0.025 |
| ptprfb | 0.041 | islr2 | -0.025 |
| her9 | 0.040 | stmn2b | -0.025 |
| scxb | 0.039 | tuba2 | -0.024 |
| ptn | 0.039 | onecut1 | -0.024 |
| CR450687.1 | 0.039 | zgc:65894 | -0.024 |
| zgc:172143 | 0.039 | inab | -0.024 |
| XLOC-040962 | 0.038 | id4 | -0.024 |
| olig4 | 0.038 | cplx2l | -0.024 |