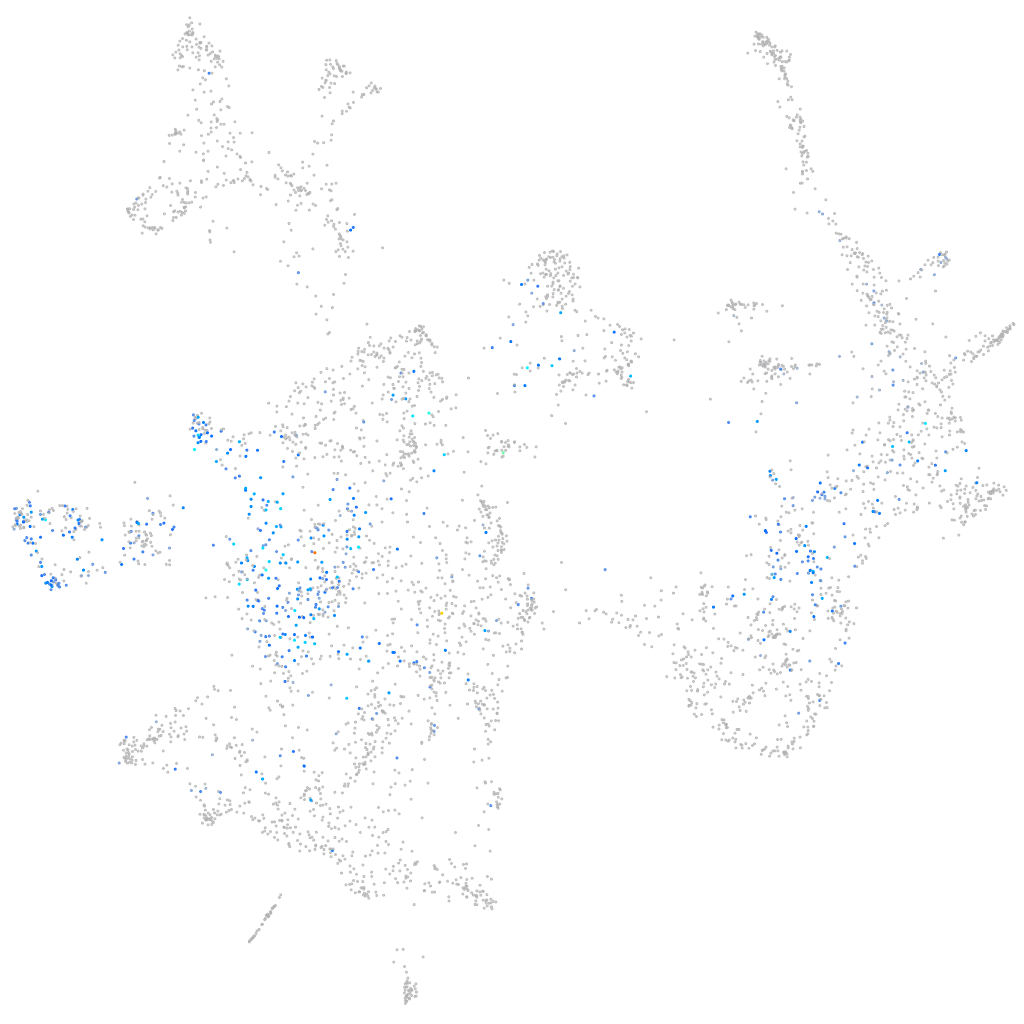
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| rrm1 | 0.472 | tob1b | -0.166 |
| pcna | 0.464 | tmem59 | -0.150 |
| rrm2 | 0.459 | COX3 | -0.149 |
| dut | 0.437 | h1f0 | -0.148 |
| slbp | 0.418 | atp2b1a | -0.144 |
| chaf1a | 0.414 | b2ml | -0.141 |
| fen1 | 0.410 | ccni | -0.137 |
| zgc:110540 | 0.407 | pvalb8 | -0.136 |
| lig1 | 0.406 | mt-co2 | -0.136 |
| CABZ01005379.1 | 0.405 | rnasekb | -0.130 |
| mibp | 0.403 | otofb | -0.129 |
| cdca5 | 0.396 | pvalb1 | -0.129 |
| dek | 0.394 | eef1da | -0.128 |
| rpa2 | 0.390 | si:dkey-16p21.8 | -0.128 |
| nasp | 0.389 | calml4a | -0.126 |
| asf1ba | 0.388 | calm1b | -0.125 |
| cks1b | 0.386 | gapdhs | -0.125 |
| zgc:110216 | 0.385 | nupr1a | -0.125 |
| ccna2 | 0.383 | atp6v1e1b | -0.125 |
| stmn1a | 0.382 | CR383676.1 | -0.123 |
| dhfr | 0.381 | socs3a | -0.123 |
| esco2 | 0.374 | gsto2 | -0.122 |
| si:dkey-6i22.5 | 0.374 | calm1a | -0.121 |
| rpa3 | 0.373 | pvalb2 | -0.121 |
| prim2 | 0.370 | kif1aa | -0.121 |
| hmgb2a | 0.363 | gpx2 | -0.121 |
| mcm7 | 0.355 | cd164l2 | -0.120 |
| mis12 | 0.355 | nptna | -0.120 |
| tubb2b | 0.354 | atp5if1b | -0.119 |
| shmt1 | 0.352 | gabarapl2 | -0.119 |
| dnajc9 | 0.351 | atp1a3b | -0.118 |
| rbbp4 | 0.348 | ip6k2a | -0.116 |
| trip13 | 0.347 | tob1a | -0.116 |
| fbxo5 | 0.345 | dnajc5b | -0.115 |
| anp32b | 0.345 | ier2b | -0.115 |