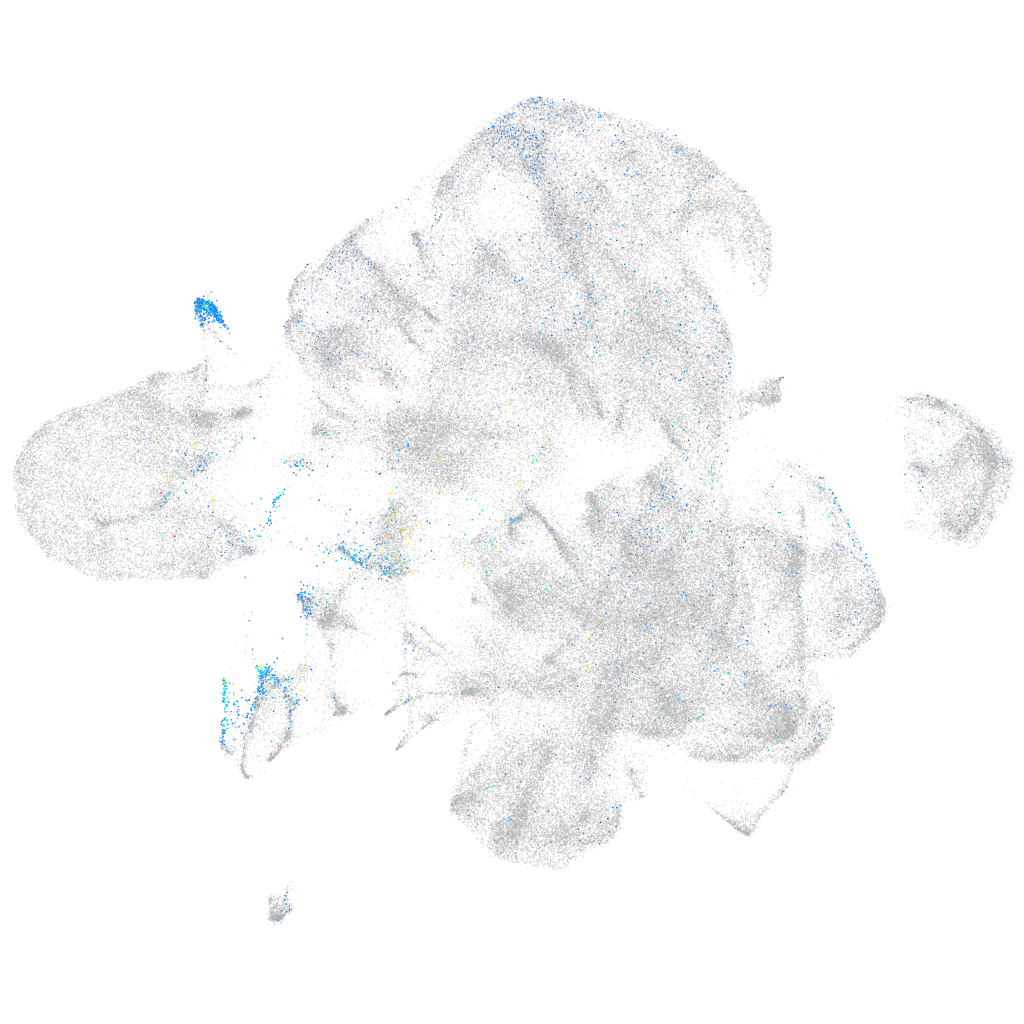
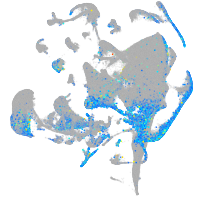
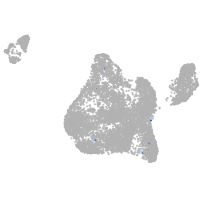
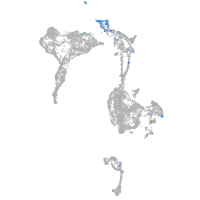
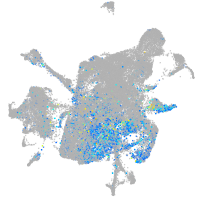
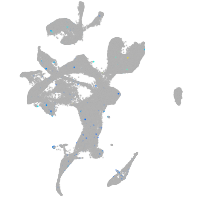
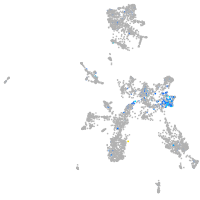
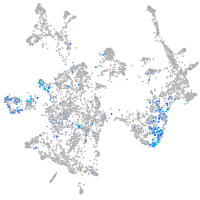
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| XLOC-000322 | 0.243 | gpm6aa | -0.072 |
| ednrab | 0.240 | nova2 | -0.065 |
| XLOC-032159 | 0.239 | tuba1c | -0.065 |
| si:dkey-165e24.1 | 0.171 | hmgb1a | -0.060 |
| inka1a | 0.165 | ckbb | -0.058 |
| snai1b | 0.150 | mdkb | -0.058 |
| XLOC-032110 | 0.142 | hmgb1b | -0.057 |
| twist1a | 0.135 | rtn1a | -0.056 |
| pmp22a | 0.129 | si:ch211-137a8.4 | -0.050 |
| crestin | 0.126 | stmn1b | -0.047 |
| pdgfra | 0.121 | gpm6ab | -0.047 |
| mfap2 | 0.117 | si:dkey-276j7.1 | -0.046 |
| six1b | 0.114 | pvalb1 | -0.045 |
| XLOC-001964 | 0.113 | gng3 | -0.045 |
| isl2b | 0.112 | zc4h2 | -0.045 |
| tuba8l3 | 0.107 | atp6v0cb | -0.044 |
| six4b | 0.105 | cspg5a | -0.044 |
| tmem119b | 0.104 | nova1 | -0.044 |
| si:ch211-11c3.9 | 0.101 | pvalb2 | -0.044 |
| dlc1 | 0.101 | rtn1b | -0.043 |
| XLOC-001603 | 0.099 | celf2 | -0.042 |
| atf3 | 0.096 | CU634008.1 | -0.041 |
| ccdc80l1 | 0.093 | gapdhs | -0.041 |
| ets1 | 0.089 | cbx1b | -0.040 |
| nid2a | 0.086 | rnasekb | -0.039 |
| cyth1b | 0.086 | hmgb3a | -0.039 |
| sox10 | 0.085 | ptmaa | -0.038 |
| mmp15b | 0.085 | pbx3b | -0.038 |
| six1a | 0.084 | CU467822.1 | -0.038 |
| junba | 0.084 | fez1 | -0.037 |
| XLOC-039121 | 0.083 | elavl4 | -0.037 |
| lin28a | 0.083 | vamp2 | -0.036 |
| nr2f5 | 0.083 | tnrc6c1 | -0.036 |
| abtb2b | 0.082 | myt1la | -0.036 |
| cxcr4b | 0.082 | ywhag2 | -0.035 |