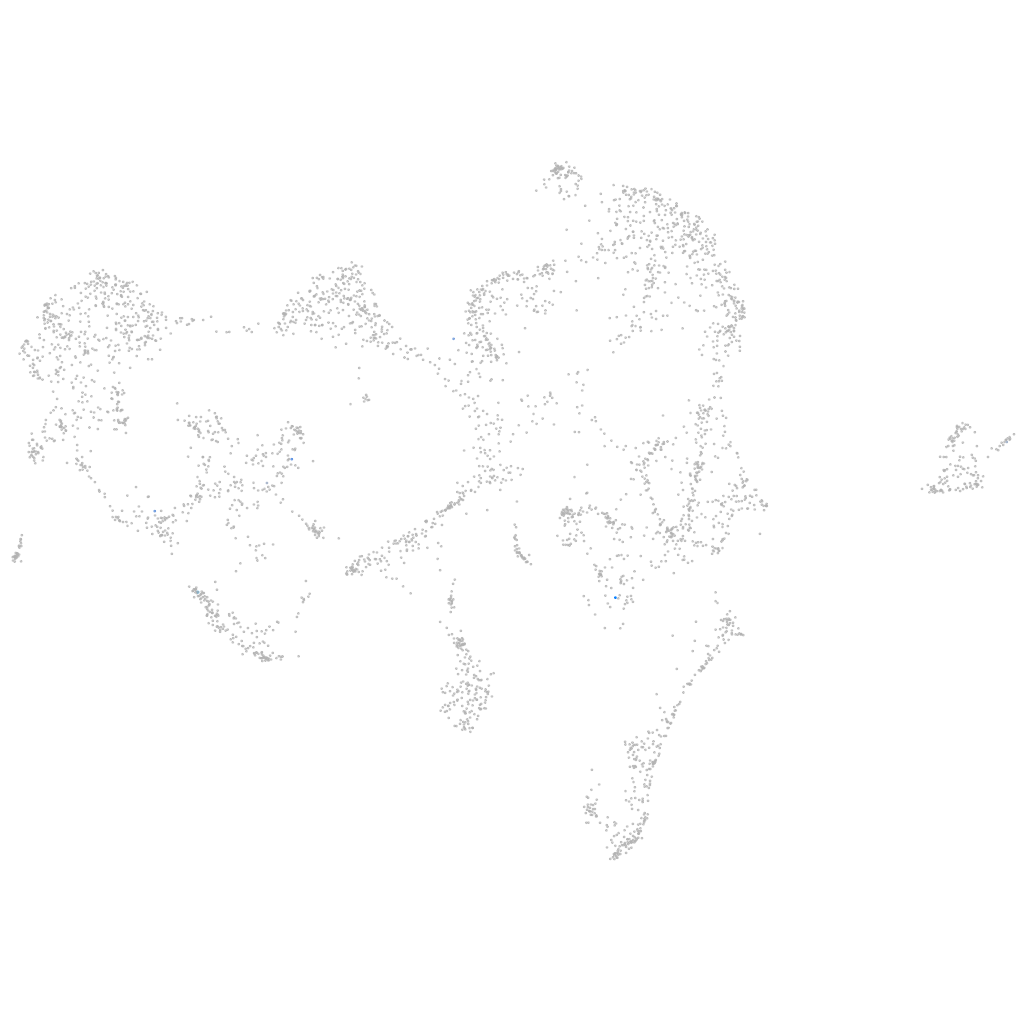
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature 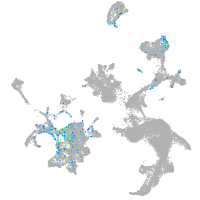 |
pronephros 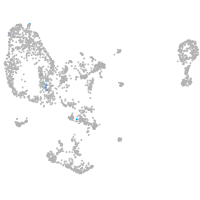 |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin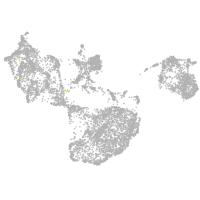 |
ionocytes / mucous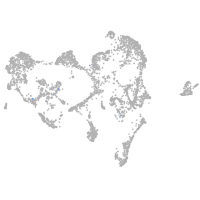 |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| XLOC-043418 | 0.572 | ier2b | -0.033 |
| veph1 | 0.458 | si:dkey-151g10.6 | -0.031 |
| LOC108180727 | 0.433 | ccdc124 | -0.030 |
| BX957328.1 | 0.414 | psmb1 | -0.029 |
| gypc | 0.378 | vcp | -0.029 |
| adgrl4 | 0.372 | cldnb | -0.028 |
| rasip1 | 0.367 | btg1 | -0.028 |
| kdrl | 0.356 | cnpy1 | -0.027 |
| aqp8a.1 | 0.339 | rps29 | -0.027 |
| shank2 | 0.321 | atf4a | -0.026 |
| kcnq4 | 0.309 | psmd7 | -0.025 |
| tmem88a | 0.303 | fam133b | -0.024 |
| LOC100333926 | 0.290 | ptmaa | -0.024 |
| aqp1a.1 | 0.283 | rpl26 | -0.024 |
| clec14a | 0.282 | hbae3 | -0.023 |
| mcamb | 0.275 | nono | -0.023 |
| ky | 0.273 | tars | -0.023 |
| FP016239.1 | 0.269 | nop58 | -0.023 |
| nipsnap1 | 0.267 | chd4a | -0.023 |
| itgae.1 | 0.262 | znf706 | -0.023 |
| esm1 | 0.255 | nutf2l | -0.023 |
| si:ch73-116o1.2 | 0.253 | prmt1 | -0.023 |
| cdh5 | 0.250 | gosr1 | -0.023 |
| flt4 | 0.248 | csnk1da | -0.022 |
| tmem54b | 0.244 | rpl29 | -0.022 |
| ecscr | 0.237 | eif2s1b | -0.022 |
| si:dkeyp-59c12.1 | 0.237 | CABZ01092156.1 | -0.021 |
| hspa12b | 0.231 | itpka | -0.021 |
| slc22a7b.1 | 0.225 | tomm40 | -0.021 |
| XLOC-005449 | 0.224 | ptmab | -0.021 |
| tnfaip2b | 0.223 | gapdhs | -0.021 |
| lmln | 0.223 | tuba8l2 | -0.021 |
| XLOC-032001 | 0.220 | si:ch73-347e22.8 | -0.021 |
| si:ch1073-268j14.1 | 0.218 | vps28 | -0.021 |
| zmp:0000001088 | 0.218 | smc1al | -0.021 |