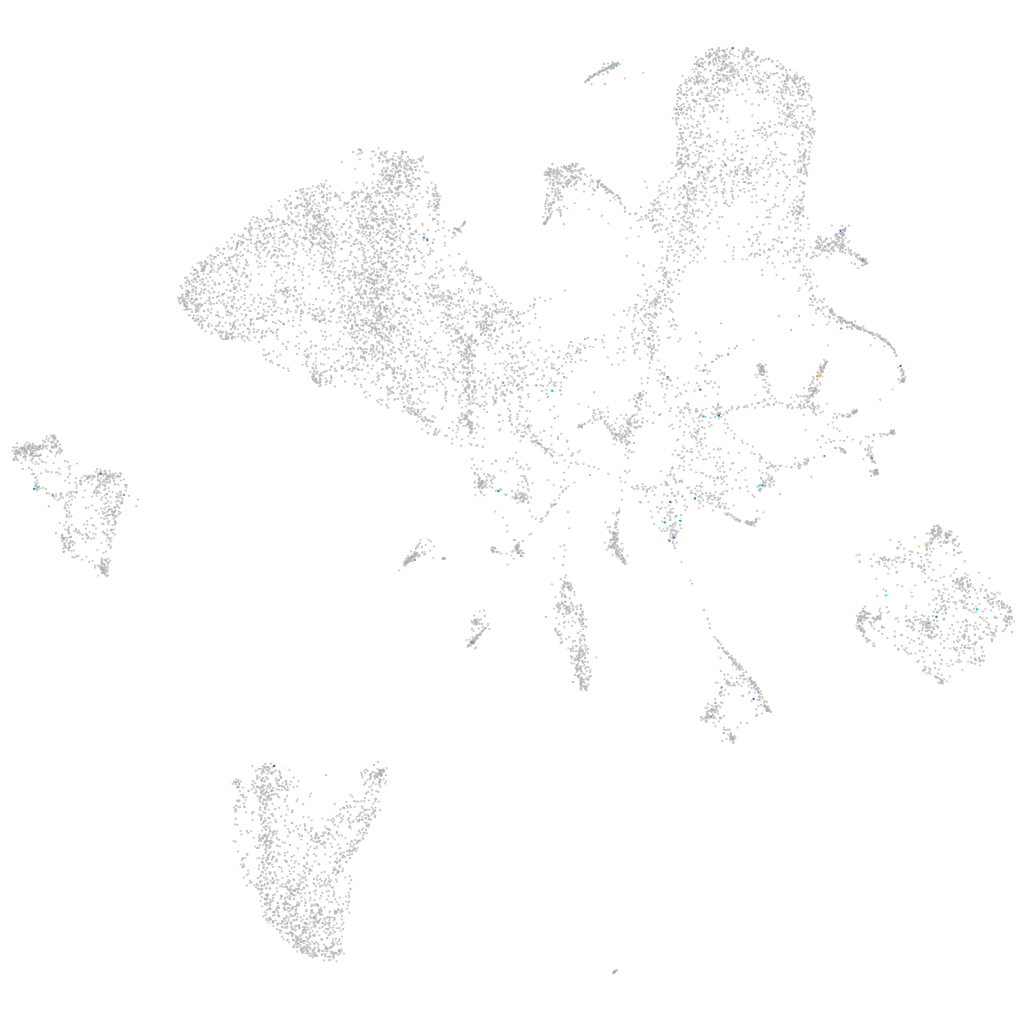
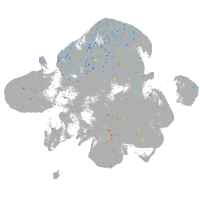
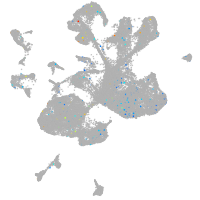
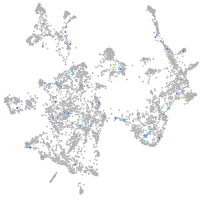
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| LO018430.1 | 0.219 | ahcy | -0.048 |
| anks6 | 0.214 | bhmt | -0.046 |
| FP325124.1 | 0.208 | gamt | -0.044 |
| mespba | 0.206 | gatm | -0.043 |
| XLOC-007006 | 0.173 | gapdh | -0.043 |
| CR936240.1 | 0.169 | apoa1b | -0.042 |
| elfn1b | 0.162 | mat1a | -0.041 |
| arhgap15 | 0.156 | dap | -0.040 |
| pnx | 0.150 | afp4 | -0.039 |
| BX908744.2 | 0.149 | apoa2 | -0.038 |
| snphb | 0.147 | agxtb | -0.038 |
| CABZ01112864.1 | 0.145 | fbp1b | -0.037 |
| si:ch211-156p11.1 | 0.144 | apoc2 | -0.036 |
| myrf | 0.141 | aqp12 | -0.036 |
| si:dkey-22i16.9 | 0.138 | grhprb | -0.036 |
| XLOC-029547 | 0.136 | atp5pf | -0.035 |
| AL954361.1 | 0.133 | zgc:123103 | -0.035 |
| XLOC-000439 | 0.132 | cox6a1 | -0.035 |
| si:ch211-232i5.3 | 0.122 | apom | -0.035 |
| si:dkeyp-44b5.4 | 0.119 | tpi1b | -0.035 |
| LOC101882853 | 0.118 | serpina1 | -0.034 |
| LOC562245 | 0.118 | eno3 | -0.034 |
| XLOC-029772 | 0.118 | mt-atp6 | -0.034 |
| zmat3 | 0.117 | tfa | -0.034 |
| syn2b | 0.116 | apoa4b.1 | -0.034 |
| CU659670.1 | 0.115 | hao1 | -0.034 |
| XLOC-020693 | 0.114 | scp2a | -0.034 |
| nppa | 0.113 | rbp2b | -0.033 |
| CR931813.1 | 0.112 | ces2 | -0.033 |
| XLOC-016178 | 0.111 | serpina1l | -0.033 |
| si:dkey-256e7.8 | 0.108 | ftcd | -0.033 |
| cplx4a | 0.105 | nipsnap3a | -0.033 |
| CU655845.1 | 0.103 | pnp4b | -0.033 |
| LOC101886167 | 0.102 | ndrg2 | -0.033 |
| crb1 | 0.101 | fgg | -0.033 |