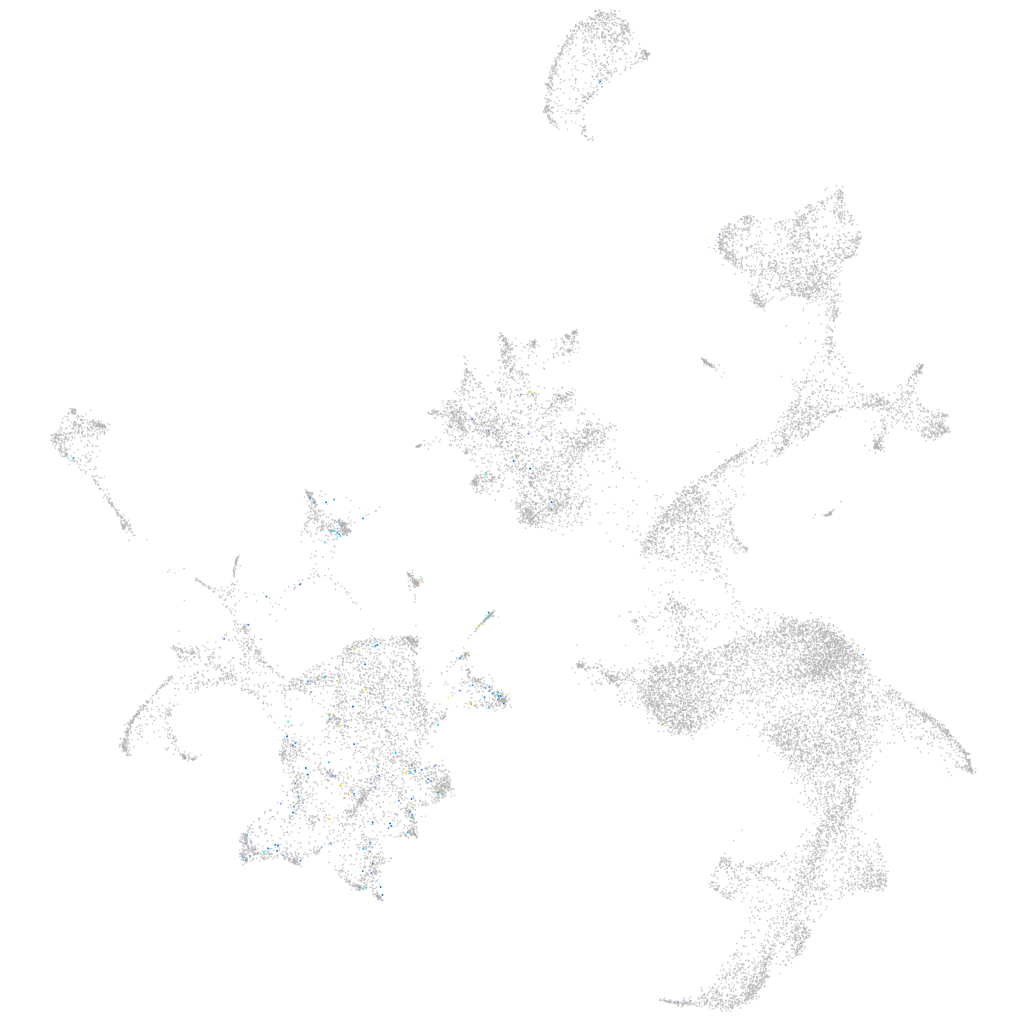
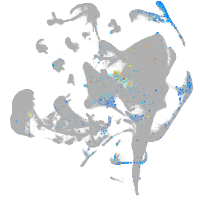
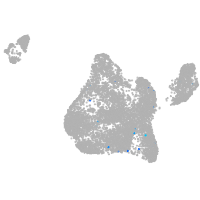
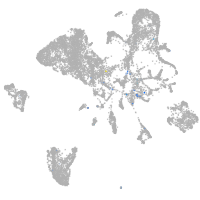
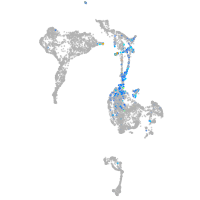
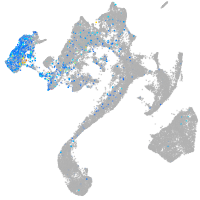
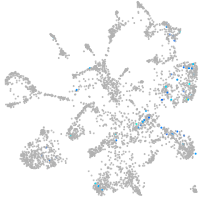
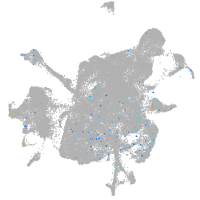
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| trim35-3 | 0.160 | prdx2 | -0.066 |
| si:dkey-250k15.9 | 0.126 | hbae1.1 | -0.065 |
| CR677513.1 | 0.126 | hbbe2 | -0.065 |
| si:ch73-242m19.1 | 0.114 | hbae1.3 | -0.065 |
| bcdin3d | 0.113 | hbae3 | -0.061 |
| CABZ01111555.1 | 0.113 | hbbe1.1 | -0.060 |
| XLOC-023345 | 0.111 | cahz | -0.059 |
| CU062454.1 | 0.109 | hbbe1.3 | -0.058 |
| LOC110437725 | 0.109 | hemgn | -0.058 |
| znf1016 | 0.109 | blvrb | -0.057 |
| LOC108179662 | 0.109 | hbbe1.2 | -0.055 |
| ecscr | 0.105 | tspo | -0.055 |
| nr5a2 | 0.104 | creg1 | -0.053 |
| cpn1 | 0.103 | slc4a1a | -0.053 |
| CABZ01081897.1 | 0.103 | fth1a | -0.052 |
| tpm4a | 0.102 | si:ch211-250g4.3 | -0.052 |
| myct1a | 0.102 | alas2 | -0.051 |
| ramp2 | 0.102 | nt5c2l1 | -0.050 |
| flt4 | 0.100 | epb41b | -0.049 |
| tuba8l3 | 0.100 | zgc:163057 | -0.049 |
| kl | 0.099 | gpx1a | -0.047 |
| gper1 | 0.099 | nmt1b | -0.044 |
| hyal2b | 0.099 | plac8l1 | -0.042 |
| vat1 | 0.097 | si:ch211-207c6.2 | -0.042 |
| rasip1 | 0.097 | tmod4 | -0.040 |
| crip2 | 0.097 | slc11a2 | -0.038 |
| marcksl1a | 0.097 | znfl2a | -0.038 |
| lamb1b | 0.097 | tfr1a | -0.038 |
| tmem88a | 0.095 | sptb | -0.037 |
| egfl7 | 0.095 | rfesd | -0.037 |
| dab2ipb | 0.094 | hdr | -0.037 |
| si:ch211-156j16.1 | 0.093 | hbae5 | -0.036 |
| krt18a.1 | 0.092 | add2 | -0.036 |
| si:ch211-33e4.2 | 0.092 | mcl1a | -0.036 |
| afap1l1b | 0.092 | klf1 | -0.036 |