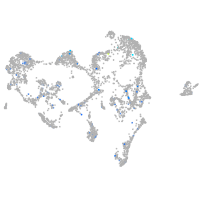
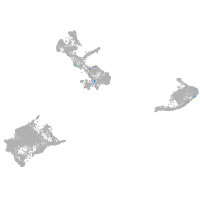
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| BX842701.1 | 0.225 | bhmt | -0.065 |
| ctbs | 0.179 | gatm | -0.059 |
| cyldb | 0.168 | gamt | -0.059 |
| pimr176 | 0.159 | apoa1b | -0.058 |
| znf1079 | 0.152 | aqp12 | -0.054 |
| LOC101886581 | 0.151 | mat1a | -0.054 |
| BX296526.2 | 0.151 | ahcy | -0.054 |
| magixb | 0.151 | gapdh | -0.052 |
| si:dkey-85n7.8 | 0.147 | grhprb | -0.052 |
| ankmy1 | 0.141 | agxtb | -0.052 |
| trim35-29 | 0.140 | apoa2 | -0.051 |
| ppfia2 | 0.138 | rbp2b | -0.050 |
| itln1 | 0.136 | serpina1l | -0.050 |
| BX548046.2 | 0.135 | serpina1 | -0.050 |
| XLOC-010592 | 0.133 | zgc:123103 | -0.050 |
| acp2 | 0.133 | agxta | -0.048 |
| si:dkeyp-51f12.3 | 0.132 | fgg | -0.048 |
| LOC101884096 | 0.130 | cfhl4 | -0.048 |
| si:ch211-207i1.2 | 0.127 | hao1 | -0.048 |
| smpx | 0.124 | pklr | -0.048 |
| barx2 | 0.124 | fetub | -0.048 |
| CR589947.2 | 0.120 | kng1 | -0.047 |
| CR847944.2 | 0.118 | ambp | -0.047 |
| FP236161.1 | 0.118 | hpda | -0.047 |
| znfl1c | 0.118 | gpd1b | -0.047 |
| LOC101885758 | 0.115 | ces2 | -0.047 |
| cyp2k31 | 0.113 | apom | -0.047 |
| ldb2b | 0.112 | pcbd1 | -0.046 |
| RF00600 | 0.111 | tfa | -0.046 |
| ftr86 | 0.108 | uox | -0.046 |
| spartb | 0.107 | LOC110437731 | -0.046 |
| LOC101882884 | 0.107 | afp4 | -0.045 |
| LOC103910264 | 0.105 | nipsnap3a | -0.045 |
| flrt2 | 0.103 | fabp10a | -0.045 |
| zgc:101583 | 0.102 | apoc1 | -0.045 |