Other cell groups


all cells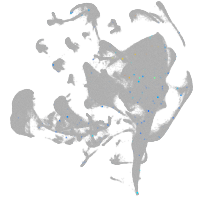 |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory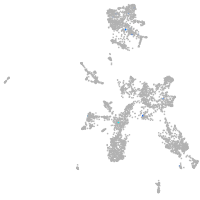 |
otic / lateral line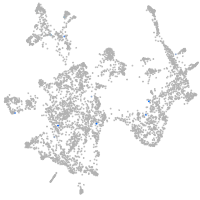 |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells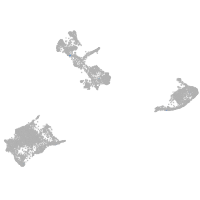 |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| cad | 0.018 | pvalb1 | -0.009 |
| cbx3a | 0.017 | cyt1 | -0.008 |
| hnrnpul1 | 0.017 | cyt1l | -0.008 |
| ptges3b | 0.017 | icn | -0.008 |
| setb | 0.017 | icn2 | -0.008 |
| tma16 | 0.017 | krt4 | -0.008 |
| apex1 | 0.016 | krtt1c19e | -0.008 |
| cct7 | 0.016 | pvalb2 | -0.008 |
| eif3ba | 0.016 | si:ch211-195b11.3 | -0.008 |
| hells | 0.016 | wu:fb18f06 | -0.008 |
| mcm6 | 0.016 | actc1b | -0.007 |
| ncl | 0.016 | anxa1a | -0.007 |
| nop2 | 0.016 | anxa1c | -0.007 |
| nop56 | 0.016 | ckap4 | -0.007 |
| npm1a | 0.016 | hrc | -0.007 |
| tcp1 | 0.016 | krt17 | -0.007 |
| abce1 | 0.015 | krt5 | -0.007 |
| bysl | 0.015 | lye | -0.007 |
| cct4 | 0.015 | mylpfa | -0.007 |
| cct5 | 0.015 | si:ch211-125o16.4 | -0.007 |
| ddx39ab | 0.015 | si:ch211-207n23.2 | -0.007 |
| ddx61 | 0.015 | si:dkey-16p21.8 | -0.007 |
| dkc1 | 0.015 | si:dkey-222n6.2 | -0.007 |
| eif3s6ip | 0.015 | si:dkey-247k7.2 | -0.007 |
| eif4a1a | 0.015 | tcnbb | -0.007 |
| etf1b | 0.015 | tmem176l.4 | -0.007 |
| mcm2 | 0.015 | wu:fa03e10 | -0.007 |
| mcm4 | 0.015 | zgc:193505 | -0.007 |
| nop58 | 0.015 | abca12 | -0.006 |
| prrc2c | 0.015 | abcb5 | -0.006 |
| ranbp1 | 0.015 | aep1 | -0.006 |
| si:dkey-102m7.3 | 0.015 | agr1 | -0.006 |
| si:dkey-23i12.5 | 0.015 | anxa1b | -0.006 |
| snrpg | 0.015 | apoa2 | -0.006 |
| snu13b | 0.015 | apodb | -0.006 |




