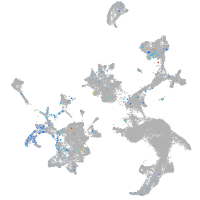
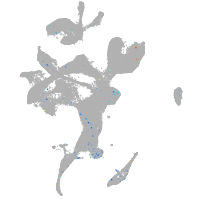
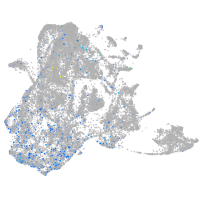
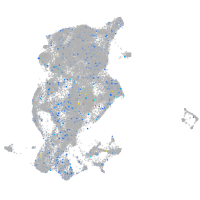
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| XLOC-032526 | 0.067 | elavl3 | -0.044 |
| CABZ01075068.1 | 0.065 | rtn1a | -0.042 |
| npm1a | 0.059 | ckbb | -0.041 |
| paics | 0.059 | tuba1c | -0.038 |
| eif4ebp3l | 0.057 | gpm6aa | -0.038 |
| nhp2 | 0.057 | tmsb | -0.038 |
| snu13b | 0.057 | myt1b | -0.036 |
| selenoh | 0.057 | pvalb1 | -0.036 |
| XLOC-039121 | 0.056 | stmn1b | -0.036 |
| cnbpa | 0.056 | pvalb2 | -0.035 |
| atf3 | 0.055 | COX3 | -0.034 |
| nop10 | 0.054 | nova2 | -0.032 |
| id1 | 0.054 | hmgb3a | -0.032 |
| nutf2l | 0.053 | hbbe1.3 | -0.032 |
| BX927258.1 | 0.053 | ptmaa | -0.032 |
| cdca7b | 0.053 | CU634008.1 | -0.031 |
| FO904898.1 | 0.052 | myt1a | -0.030 |
| emg1 | 0.052 | hbae3 | -0.030 |
| nop58 | 0.052 | gpm6ab | -0.029 |
| mcm6 | 0.052 | rnasekb | -0.028 |
| mcm3 | 0.052 | tuba1a | -0.028 |
| rsl1d1 | 0.051 | CU467822.1 | -0.027 |
| pcna | 0.051 | scrt2 | -0.027 |
| odc1 | 0.051 | chd4a | -0.027 |
| ranbp1 | 0.051 | hbae1.1 | -0.027 |
| junba | 0.051 | ube2d4 | -0.027 |
| adi1 | 0.051 | actc1b | -0.027 |
| cdca7a | 0.051 | elavl4 | -0.026 |
| dkc1 | 0.050 | gng3 | -0.026 |
| mcm5 | 0.050 | fez1 | -0.026 |
| aldob | 0.049 | atp6v0cb | -0.026 |
| ctps1a | 0.049 | celf3a | -0.026 |
| prps1a | 0.049 | sncb | -0.026 |
| sdc4 | 0.049 | mt-co2 | -0.025 |
| cad | 0.049 | fam168a | -0.025 |