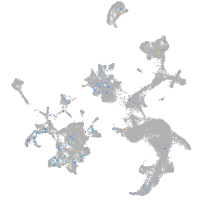
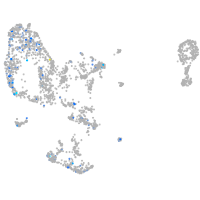
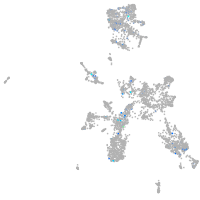
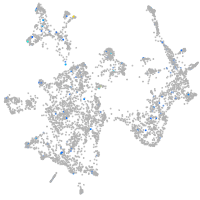
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| elavl3 | 0.089 | hspb1 | -0.035 |
| myt1b | 0.076 | npm1a | -0.034 |
| nova2 | 0.073 | dkc1 | -0.033 |
| stmn1b | 0.071 | eif4ebp3l | -0.033 |
| tubb5 | 0.071 | nop58 | -0.033 |
| myt1a | 0.070 | banf1 | -0.032 |
| tmsb | 0.070 | nop56 | -0.031 |
| gng2 | 0.069 | anp32b | -0.030 |
| rtn1a | 0.069 | fbl | -0.030 |
| tmeff1b | 0.068 | rsl1d1 | -0.030 |
| tuba1c | 0.068 | aldob | -0.029 |
| dpysl3 | 0.067 | chaf1a | -0.029 |
| nhlh2 | 0.067 | lig1 | -0.029 |
| ywhah | 0.066 | pcna | -0.029 |
| tuba1a | 0.065 | si:ch211-152c2.3 | -0.029 |
| dpysl2b | 0.064 | snu13b | -0.029 |
| hmgb3a | 0.062 | apoeb | -0.028 |
| scrt1a | 0.062 | ccnd1 | -0.028 |
| si:dkey-276j7.1 | 0.062 | ccng1 | -0.028 |
| celf3a | 0.061 | nop10 | -0.028 |
| gng3 | 0.061 | polr3gla | -0.028 |
| jpt1b | 0.060 | apoc1 | -0.027 |
| scrt2 | 0.060 | cdca7a | -0.027 |
| fez1 | 0.059 | dek | -0.027 |
| si:ch73-386h18.1 | 0.059 | gar1 | -0.027 |
| stx1b | 0.059 | gnl3 | -0.027 |
| zc4h2 | 0.059 | id1 | -0.027 |
| stxbp1a | 0.059 | mki67 | -0.027 |
| elavl4 | 0.056 | nhp2 | -0.027 |
| gpm6aa | 0.056 | nop2 | -0.027 |
| mllt11 | 0.056 | stm | -0.027 |
| vamp2 | 0.056 | stmn1a | -0.027 |
| zfhx3 | 0.056 | ddx18 | -0.026 |
| adam22 | 0.055 | hdlbpa | -0.026 |
| gdf11 | 0.055 | pou5f3 | -0.026 |