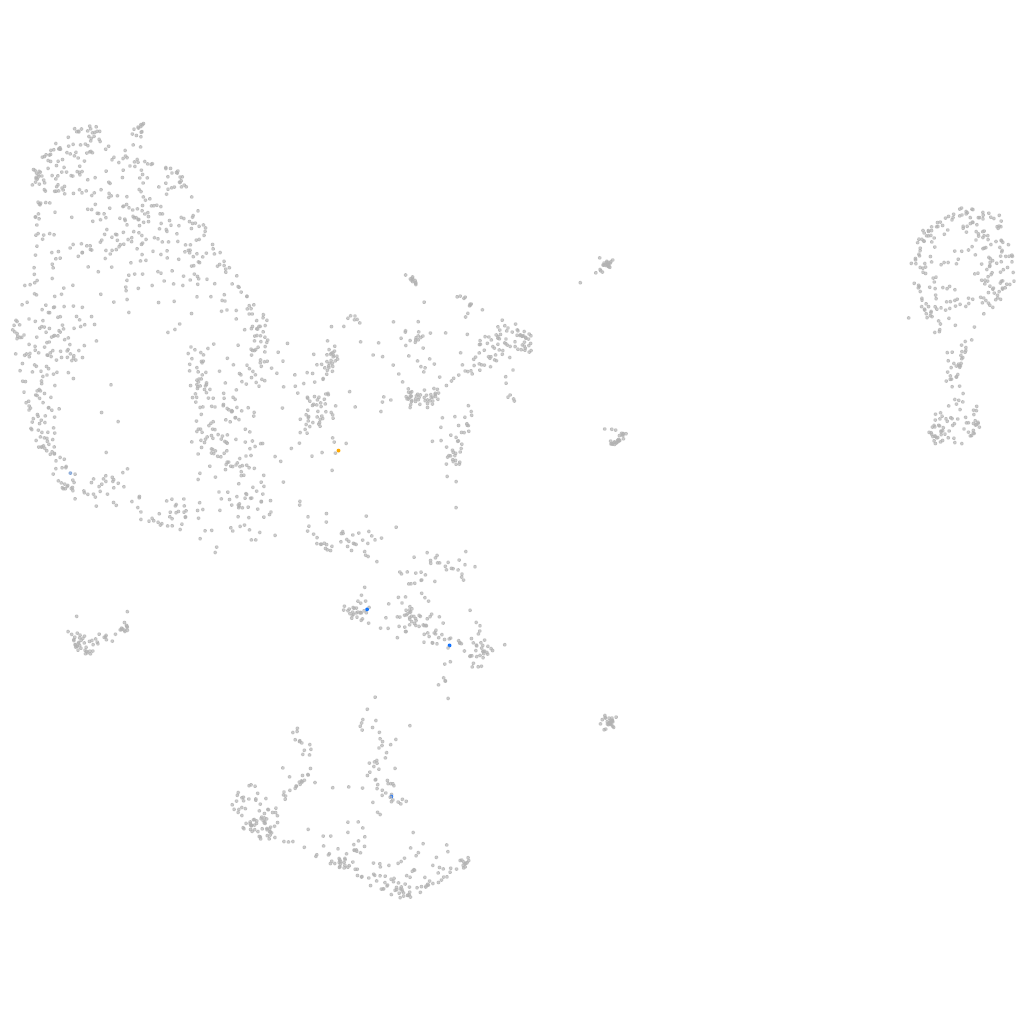
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| CR352226.2 | 0.912 | mt-co2 | -0.085 |
| mhc2dbb | 0.844 | rps6 | -0.077 |
| lrata | 0.572 | eef2b | -0.071 |
| zmp:0000000760 | 0.544 | COX3 | -0.070 |
| pld1b | 0.512 | rpl27a | -0.068 |
| atp1a2a | 0.502 | pabpc1a | -0.065 |
| tmem38a | 0.396 | NC-002333.4 | -0.062 |
| si:dkey-269i1.3 | 0.361 | cox7c | -0.058 |
| si:ch211-79l17.1 | 0.357 | mt-co1 | -0.055 |
| ncapg2 | 0.316 | rpl6 | -0.054 |
| mvda | 0.304 | nme2b.1 | -0.053 |
| si:ch1073-296i8.2 | 0.274 | rps18 | -0.053 |
| fadd | 0.272 | mt-atp6 | -0.052 |
| si:dkeyp-110g5.4 | 0.271 | ppiaa | -0.049 |
| pde9a | 0.266 | sec61g | -0.049 |
| map3k15 | 0.262 | rpl17 | -0.047 |
| CR925719.1 | 0.247 | fthl27 | -0.046 |
| si:ch211-102c2.7 | 0.242 | zgc:193541 | -0.045 |
| trim35-7 | 0.240 | fth1a | -0.045 |
| gig2g | 0.240 | cox6c | -0.045 |
| LOC101885934 | 0.240 | pnrc2 | -0.041 |
| haus6 | 0.239 | mt-nd1 | -0.041 |
| mcm10 | 0.234 | mt-nd4 | -0.040 |
| sytl1 | 0.231 | mt-nd3 | -0.037 |
| trip13 | 0.231 | ndufb8 | -0.037 |
| chek1 | 0.229 | sod1 | -0.036 |
| hs6st1a | 0.229 | romo1 | -0.035 |
| serpinb14 | 0.228 | eno3 | -0.035 |
| mcat | 0.226 | aldob | -0.035 |
| dbf4b | 0.226 | ppib | -0.035 |
| ches1 | 0.221 | mt-nd2 | -0.034 |
| enc2 | 0.221 | ldhba | -0.034 |
| rad18 | 0.219 | NC-002333.17 | -0.034 |
| CABZ01088271.1 | 0.218 | si:ch1073-325m22.2 | -0.033 |
| actr8 | 0.217 | ccni | -0.033 |