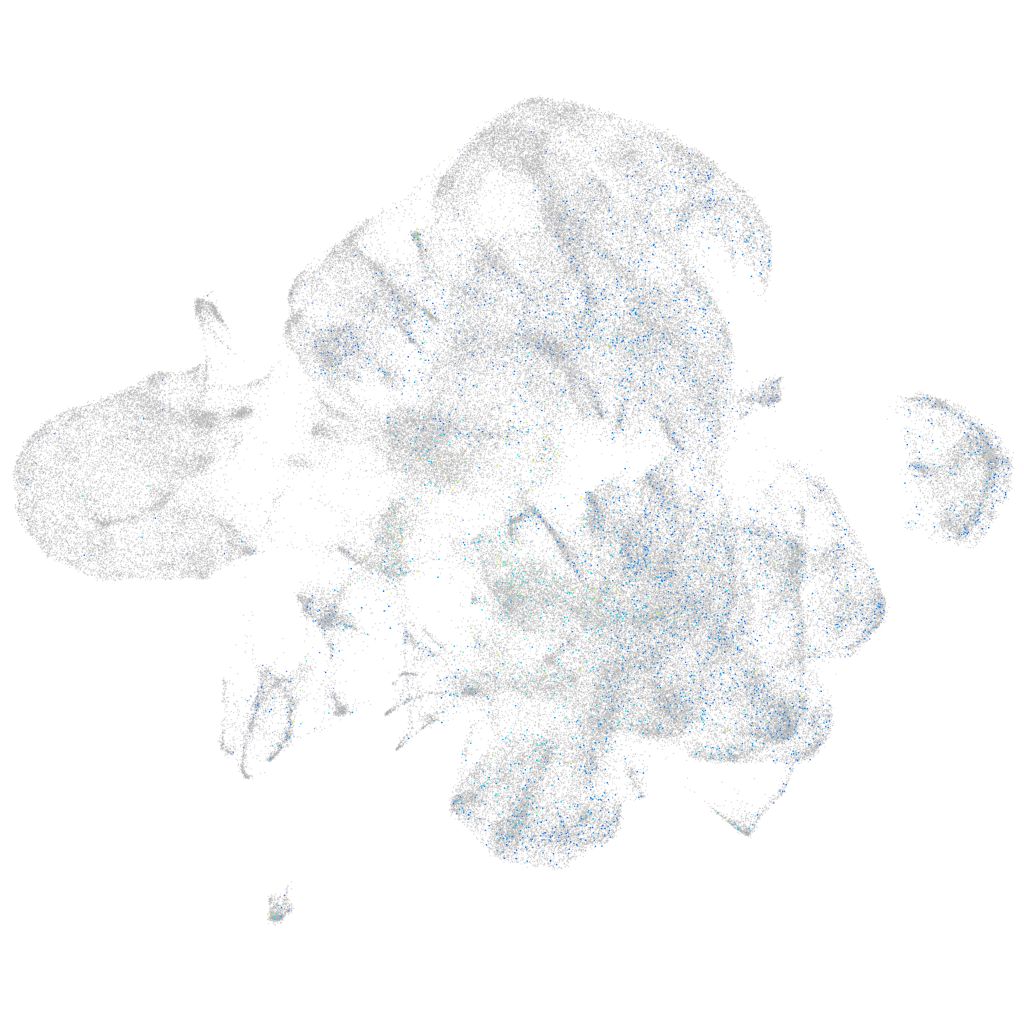
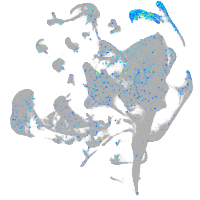
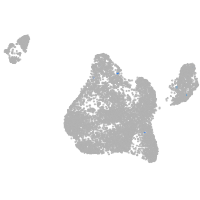
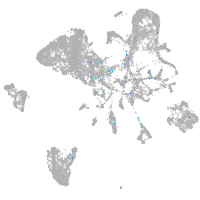
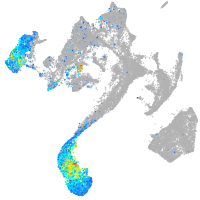
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| hbbe1.3 | 0.080 | npm1a | -0.059 |
| actc1b | 0.079 | nop58 | -0.057 |
| pvalb2 | 0.077 | hspb1 | -0.055 |
| pvalb1 | 0.076 | snu13b | -0.054 |
| hbae3 | 0.069 | dkc1 | -0.053 |
| acta1b | 0.068 | rsl1d1 | -0.053 |
| mylpfa | 0.066 | nop56 | -0.053 |
| hbae1.1 | 0.065 | anp32b | -0.053 |
| krt4 | 0.060 | fbl | -0.052 |
| ckma | 0.058 | pcna | -0.052 |
| myhz1.1 | 0.057 | hmga1a | -0.051 |
| mylz3 | 0.053 | mcm6 | -0.051 |
| cyt1l | 0.053 | hmgb2a | -0.050 |
| tnni2a.4 | 0.053 | banf1 | -0.050 |
| atp2a1 | 0.051 | aldob | -0.049 |
| cyt1 | 0.049 | nop2 | -0.049 |
| CR383676.1 | 0.048 | lig1 | -0.049 |
| ptmaa | 0.048 | ranbp1 | -0.048 |
| ttn.1 | 0.048 | ncl | -0.048 |
| gpm6aa | 0.048 | selenoh | -0.048 |
| RF00397 | 0.047 | nop10 | -0.048 |
| ckbb | 0.047 | nhp2 | -0.047 |
| gpm6ab | 0.047 | chaf1a | -0.047 |
| COX3 | 0.047 | ebna1bp2 | -0.046 |
| rtn1a | 0.047 | anp32a | -0.046 |
| hbbe1.1 | 0.047 | eif5a2 | -0.046 |
| tnnt3b | 0.045 | nasp | -0.046 |
| myhz1.3 | 0.045 | cdca7b | -0.046 |
| myl1 | 0.045 | snrpb | -0.046 |
| mt-co2 | 0.044 | gar1 | -0.045 |
| ttn.2 | 0.044 | serbp1a | -0.045 |
| mylpfb | 0.044 | si:ch211-217k17.7 | -0.045 |
| hbbe1.2 | 0.043 | eif4ebp3l | -0.044 |
| tpma | 0.043 | ccnd1 | -0.044 |
| tuba1c | 0.043 | gnl3 | -0.044 |