Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia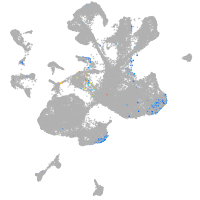 |
eye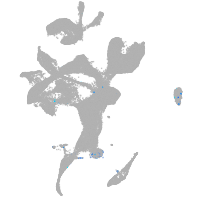 |
taste / olfactory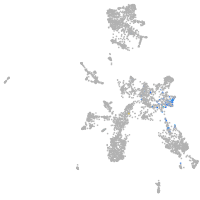 |
otic / lateral line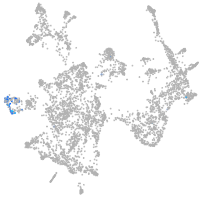 |
epidermis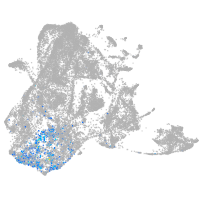 |
periderm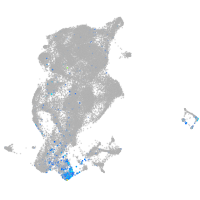 |
fin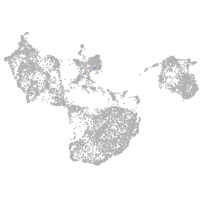 |
ionocytes / mucous |
pigment cells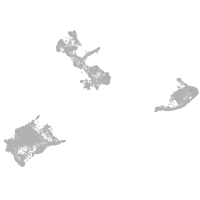 |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| XLOC-032526 | 0.415 | gpm6aa | -0.147 |
| BX927258.1 | 0.331 | rtn1a | -0.138 |
| XLOC-039121 | 0.287 | elavl3 | -0.137 |
| XLOC-026578 | 0.254 | stmn1b | -0.133 |
| ptgdsb.1 | 0.251 | ptmaa | -0.131 |
| XLOC-001603 | 0.251 | tuba1c | -0.129 |
| hspb1 | 0.248 | ckbb | -0.128 |
| ubxn8 | 0.218 | tmsb | -0.124 |
| npm1a | 0.216 | gpm6ab | -0.114 |
| snu13b | 0.204 | pvalb1 | -0.113 |
| nhp2 | 0.202 | pvalb2 | -0.112 |
| nop58 | 0.198 | CR383676.1 | -0.112 |
| dkc1 | 0.194 | actc1b | -0.109 |
| paics | 0.193 | hbbe1.3 | -0.107 |
| CABZ01075068.1 | 0.185 | gng3 | -0.104 |
| mcm6 | 0.183 | gapdhs | -0.103 |
| nop56 | 0.183 | rnasekb | -0.102 |
| fbl | 0.183 | celf2 | -0.100 |
| emg1 | 0.180 | sncb | -0.100 |
| nop10 | 0.179 | atp6v0cb | -0.100 |
| cad | 0.179 | COX3 | -0.098 |
| ctps1a | 0.176 | hbae3 | -0.098 |
| nop2 | 0.172 | ccni | -0.097 |
| mcm5 | 0.170 | elavl4 | -0.095 |
| mcm3 | 0.170 | rtn1b | -0.094 |
| cdca7b | 0.169 | vamp2 | -0.093 |
| ptges3b | 0.169 | marcksl1a | -0.092 |
| ranbp1 | 0.167 | si:dkeyp-75h12.5 | -0.091 |
| si:ch211-217k17.7 | 0.167 | myt1b | -0.091 |
| rpl7l1 | 0.167 | hbae1.1 | -0.091 |
| nip7 | 0.165 | fez1 | -0.091 |
| XLOC-020797 | 0.164 | hmgb3a | -0.090 |
| cnbpa | 0.164 | kdm6bb | -0.088 |
| si:ch211-132b12.1 | 0.164 | atp6v1e1b | -0.088 |
| si:ch211-226l4.6 | 0.163 | ywhag2 | -0.088 |




