Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle 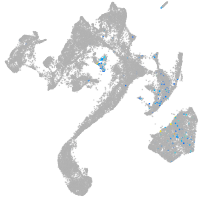 |
mural cells / non-skeletal muscle 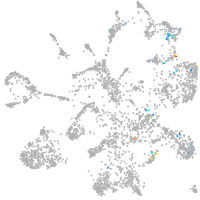 |
mesenchyme 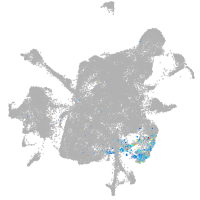 |
hematopoietic / vasculature 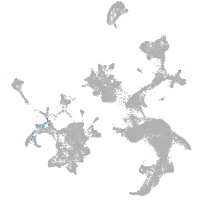 |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| fbn2a.1 | 0.219 | actc1b | -0.074 |
| hand2 | 0.183 | eef1da | -0.069 |
| tmem88b | 0.157 | fabp3 | -0.068 |
| XLOC-028299 | 0.152 | ttn.1 | -0.067 |
| gata5 | 0.145 | atp5meb | -0.066 |
| gata6 | 0.136 | ttn.2 | -0.064 |
| crb2a | 0.126 | ak1 | -0.063 |
| fgf10b | 0.125 | pabpc4 | -0.063 |
| cdh13 | 0.124 | vwde | -0.062 |
| ssuh2.2 | 0.123 | klhl41b | -0.061 |
| pcdh10a | 0.123 | tmem38a | -0.061 |
| BX469925.3 | 0.117 | atp2a1 | -0.060 |
| BX005396.3 | 0.113 | aldoab | -0.059 |
| apoeb | 0.105 | ckmb | -0.059 |
| pak6b | 0.103 | ckma | -0.059 |
| wu:fb97g03 | 0.103 | eno1a | -0.059 |
| ftr82 | 0.103 | atp5if1b | -0.059 |
| cfd | 0.099 | mdh2 | -0.057 |
| LOC108180265 | 0.098 | vdac3 | -0.057 |
| znfl2a | 0.097 | tnnc2 | -0.057 |
| pcolcea | 0.095 | mylpfa | -0.057 |
| si:dkey-28d5.13 | 0.093 | gatm | -0.056 |
| si:dkey-12j5.1 | 0.093 | mybphb | -0.056 |
| FRMD1 | 0.093 | tpma | -0.056 |
| cracr2b | 0.092 | gamt | -0.055 |
| jam2b | 0.091 | srl | -0.055 |
| bambia | 0.090 | zgc:92518 | -0.054 |
| tbx2a | 0.089 | acta1b | -0.054 |
| pth2r | 0.089 | CABZ01078594.1 | -0.054 |
| cdx4 | 0.089 | bhmt | -0.054 |
| vox | 0.089 | desma | -0.054 |
| tmem108 | 0.089 | neb | -0.054 |
| osr1 | 0.089 | myl1 | -0.053 |
| sema5bb | 0.088 | nme2b.2 | -0.053 |
| cx43.4 | 0.088 | cav3 | -0.053 |




