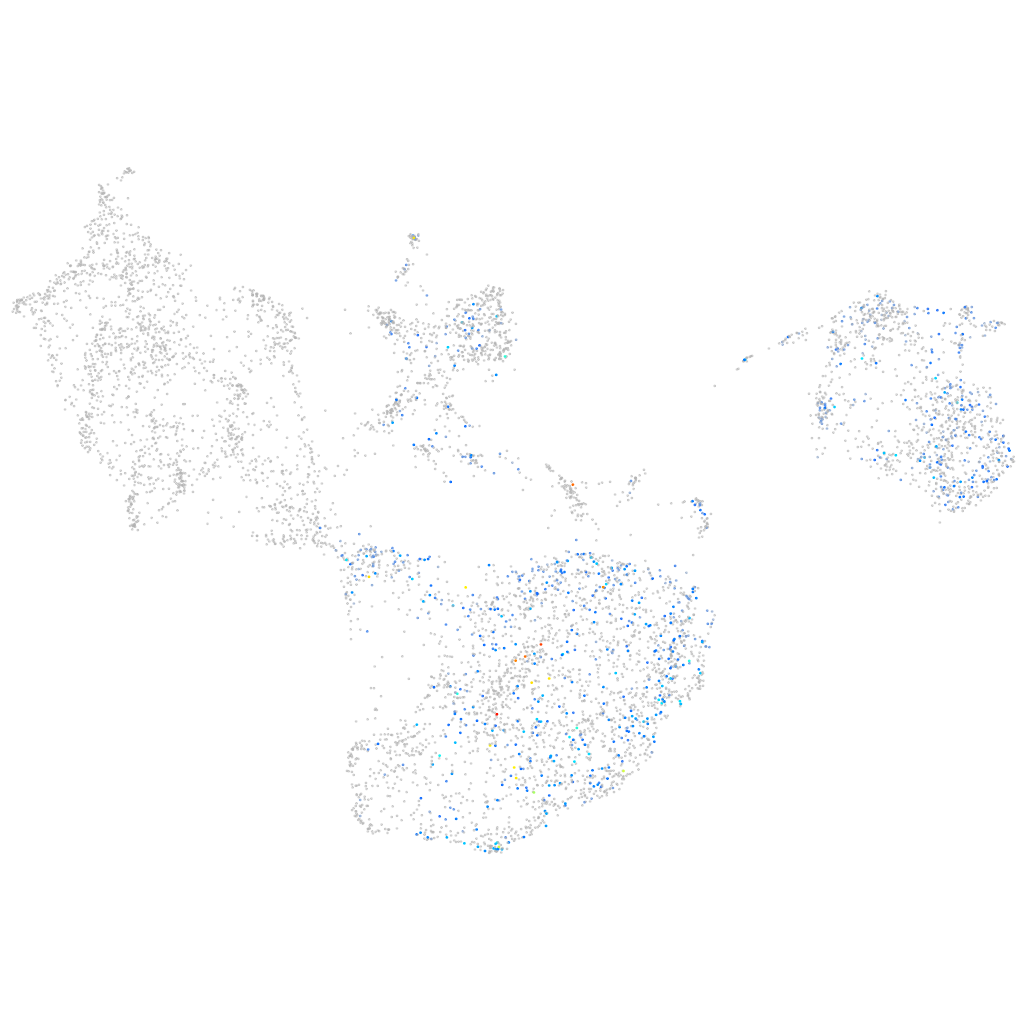
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| cotl1 | 0.245 | c1qtnf5 | -0.177 |
| cldn1 | 0.244 | bhmt | -0.167 |
| tmsb1 | 0.242 | fbln1 | -0.161 |
| cldni | 0.238 | hgd | -0.150 |
| pfn1 | 0.233 | cfl1 | -0.150 |
| cfl1l | 0.232 | cd82a | -0.148 |
| si:rp71-77l1.1 | 0.231 | ttc36 | -0.146 |
| tagln2 | 0.224 | twist1a | -0.144 |
| epcam | 0.223 | tmem119b | -0.142 |
| ecrg4b | 0.223 | pcbd1 | -0.141 |
| spaca4l | 0.223 | qdpra | -0.140 |
| thy1 | 0.221 | prrx1a | -0.138 |
| anxa1a | 0.219 | prrx1b | -0.137 |
| arpc5b | 0.217 | ptx3a | -0.136 |
| myh9a | 0.216 | hpdb | -0.134 |
| zgc:101810 | 0.216 | mfap2 | -0.133 |
| itm2bb | 0.215 | marcksl1a | -0.132 |
| arhgdig | 0.214 | fah | -0.130 |
| s100a10b | 0.213 | cd81a | -0.128 |
| si:dkey-102c8.3 | 0.212 | gpc1a | -0.127 |
| plecb | 0.212 | calr | -0.127 |
| cyt1 | 0.211 | crabp2a | -0.127 |
| zgc:92380 | 0.209 | mdh1aa | -0.125 |
| krt4 | 0.209 | msna | -0.123 |
| rbp4 | 0.208 | ntd5 | -0.123 |
| col18a1a | 0.208 | hoxa13b | -0.121 |
| apoeb | 0.207 | mxra5b | -0.120 |
| tmem238a | 0.207 | pah | -0.120 |
| igfbp2a | 0.206 | pmp22a | -0.119 |
| tmsb4x | 0.206 | tuba8l3 | -0.119 |
| rac2 | 0.205 | reck | -0.118 |
| zgc:86896 | 0.205 | twist1b | -0.117 |
| sult2st1 | 0.205 | slc16a10 | -0.116 |
| hbegfa | 0.205 | stox1 | -0.116 |
| bcam | 0.204 | gulp1a | -0.116 |