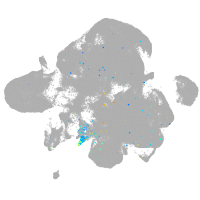
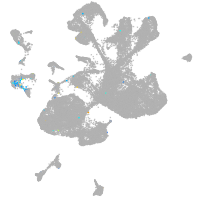
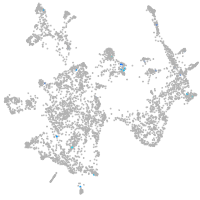
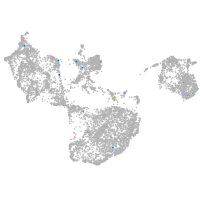
vitronectin b
ZFIN 
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| loxl5b | 0.292 | ptmab | -0.047 |
| col9a1b | 0.192 | hnrnpaba | -0.046 |
| col2a1b | 0.185 | cirbpa | -0.046 |
| optc | 0.179 | si:ch211-288g17.3 | -0.044 |
| col9a3 | 0.177 | tubb2b | -0.043 |
| hjv | 0.176 | marcksb | -0.042 |
| cers3b | 0.173 | seta | -0.040 |
| slit1b | 0.170 | nova2 | -0.040 |
| arxa | 0.162 | hmgb1b | -0.039 |
| si:dkey-16p6.4 | 0.158 | hmgn2 | -0.039 |
| clu | 0.157 | hmgb3a | -0.039 |
| shhb | 0.156 | ptmaa | -0.038 |
| col8a1a | 0.154 | hmgb1a | -0.037 |
| myl9a | 0.152 | ddx39ab | -0.037 |
| lcn15 | 0.152 | hnrnpa0b | -0.036 |
| col2a1a | 0.150 | si:dkey-56m19.5 | -0.036 |
| col9a2 | 0.147 | hdac1 | -0.035 |
| col5a1 | 0.145 | khdrbs1a | -0.035 |
| npr3 | 0.140 | snrpd1 | -0.035 |
| vasna | 0.139 | syncrip | -0.034 |
| krt94 | 0.137 | hmgb2b | -0.034 |
| aplnr2 | 0.136 | tma7 | -0.034 |
| lgals2a | 0.135 | snrpf | -0.034 |
| BX908782.3 | 0.134 | hnrnpabb | -0.033 |
| FP102018.1 | 0.134 | apex1 | -0.033 |
| cnmd | 0.131 | sox11b | -0.033 |
| sulf1 | 0.130 | elavl3 | -0.033 |
| spon1b | 0.129 | mdkb | -0.033 |
| CU651657.1 | 0.129 | chd4a | -0.033 |
| ntd5 | 0.128 | taf15 | -0.032 |
| ctgfa | 0.126 | snrpe | -0.032 |
| oc90 | 0.126 | psmb7 | -0.032 |
| scospondin | 0.126 | setb | -0.032 |
| shha | 0.124 | tmsb4x | -0.031 |
| emp2 | 0.121 | nono | -0.031 |