vacuolar protein sorting 4 homolog B
ZFIN 
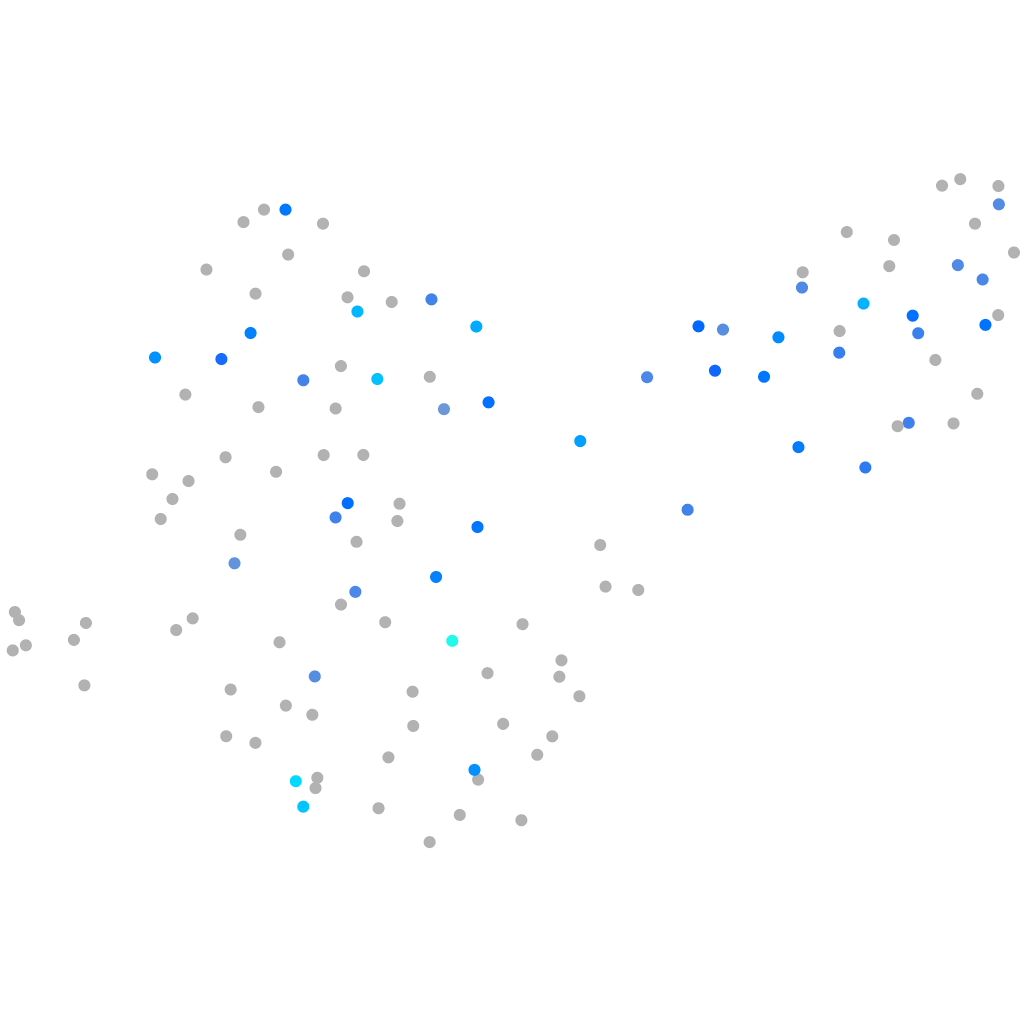
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| gchfr | 0.493 | h1m | -0.415 |
| CT956002.2 | 0.481 | cd2bp2 | -0.410 |
| ndufa3 | 0.475 | hnrnpa1b | -0.385 |
| cks2 | 0.459 | nucks1a | -0.353 |
| pbrm1 | 0.458 | si:ch73-281n10.2 | -0.340 |
| abtb1 | 0.458 | cxcr4b | -0.337 |
| flna | 0.457 | hnrnpaba | -0.330 |
| carm1l | 0.452 | stm | -0.329 |
| CU929391.1 | 0.452 | csde1 | -0.328 |
| psmb8a | 0.447 | hnrnph1l | -0.328 |
| si:dkeyp-55f12.3 | 0.442 | faf2 | -0.328 |
| map3k5 | 0.439 | sltm | -0.327 |
| icn2 | 0.437 | ctsla | -0.325 |
| si:zfos-1192g2.3 | 0.432 | rbm7 | -0.320 |
| rpl35a | 0.430 | sde2 | -0.318 |
| LOC100149647 | 0.430 | cbx1a | -0.312 |
| kank2 | 0.429 | srrt | -0.305 |
| cox7c | 0.427 | acin1a | -0.304 |
| cox8b | 0.424 | metrnla | -0.302 |
| zgc:63470 | 0.423 | avd | -0.294 |
| exd1 | 0.422 | gtf2f1 | -0.293 |
| spint1b | 0.419 | syncrip | -0.292 |
| gins2 | 0.418 | si:dkey-208k4.2 | -0.292 |
| smarca1 | 0.418 | tbx16 | -0.289 |
| rpl27 | 0.417 | zgc:112356 | -0.289 |
| sp100.2 | 0.417 | zc3h13 | -0.289 |
| FQ976913.1 | 0.417 | rcc1 | -0.284 |
| gstm.3 | 0.413 | hmgb2a | -0.284 |
| rpl26 | 0.412 | psme3 | -0.283 |
| nrp2a | 0.412 | syncripl | -0.282 |
| dele1 | 0.412 | ncl | -0.281 |
| mbnl3 | 0.411 | gar1 | -0.281 |
| tdrkh | 0.410 | ccna2 | -0.279 |
| rhbdl3 | 0.407 | ca15b | -0.277 |
| rps9 | 0.406 | ddx61 | -0.277 |