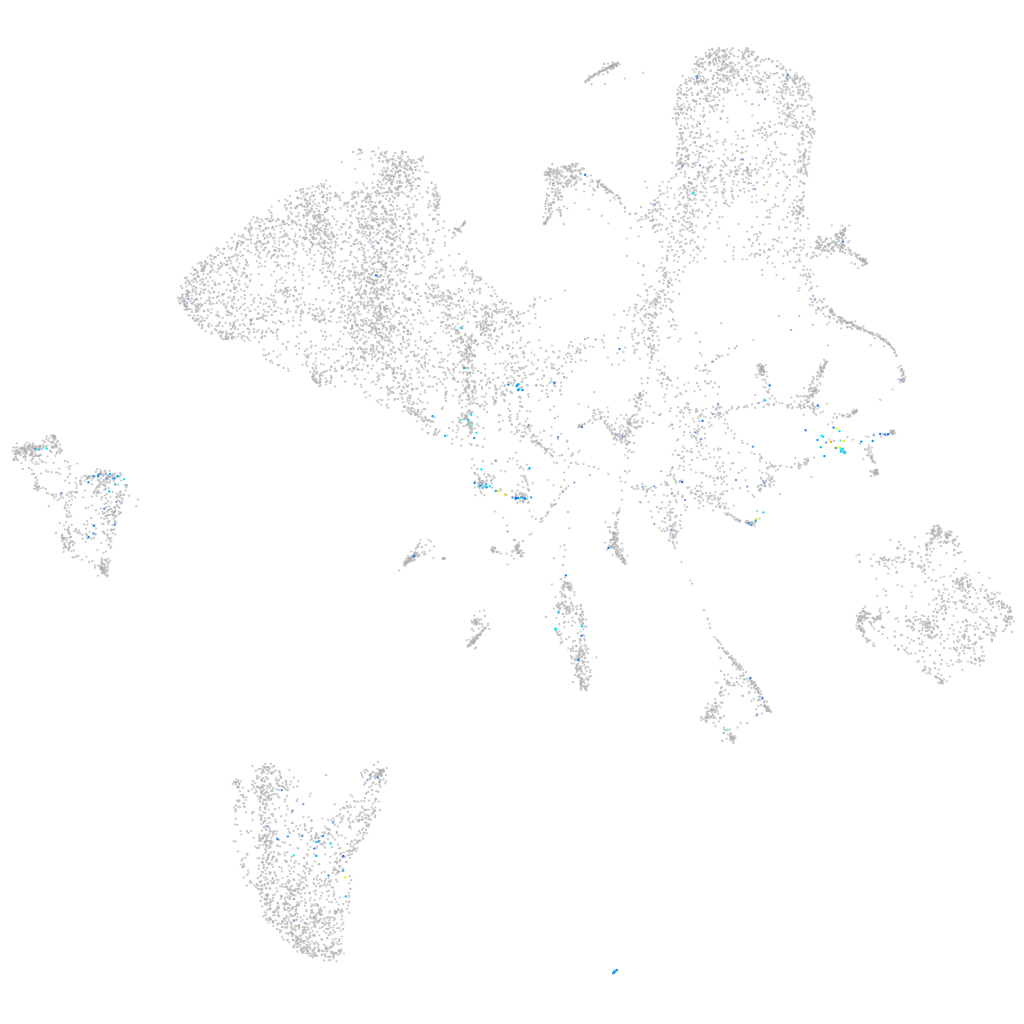
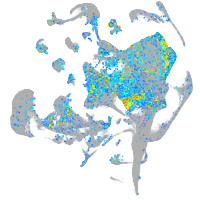
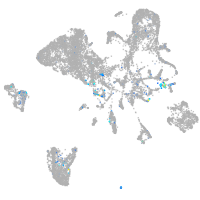
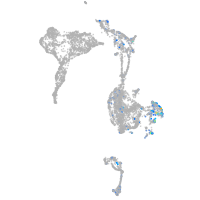
vimentin
ZFIN 
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| tg | 0.238 | aldob | -0.111 |
| slc5a5 | 0.233 | gapdh | -0.101 |
| myt1b | 0.225 | eno3 | -0.088 |
| duox2 | 0.214 | fbp1b | -0.088 |
| insm1a | 0.213 | scp2a | -0.085 |
| pygma | 0.210 | gstt1a | -0.085 |
| moxd1l | 0.201 | pklr | -0.084 |
| isl1 | 0.198 | glud1b | -0.083 |
| draxin | 0.197 | sod1 | -0.082 |
| ptx3a | 0.195 | haao | -0.081 |
| nkx2.4b | 0.186 | ahcy | -0.080 |
| myt1a | 0.186 | gstr | -0.079 |
| si:ch211-165i18.2 | 0.185 | cx32.3 | -0.079 |
| stmn1b | 0.184 | agxta | -0.078 |
| phex | 0.182 | suclg1 | -0.078 |
| ndufa4l2a | 0.180 | g6pca.2 | -0.076 |
| tpo | 0.177 | gpx1a | -0.076 |
| hapln1a | 0.176 | slco1d1 | -0.076 |
| nhlh2 | 0.176 | gatm | -0.076 |
| fam49a | 0.175 | chchd10 | -0.076 |
| dclk1a | 0.175 | apoa4b.1 | -0.075 |
| cryba2a | 0.174 | gamt | -0.075 |
| tuba1a | 0.174 | abat | -0.075 |
| pax6b | 0.173 | BX908782.3 | -0.075 |
| cnrip1a | 0.172 | rdh1 | -0.074 |
| her4.1 | 0.171 | dhrs9 | -0.074 |
| mir7a-1 | 0.170 | cat | -0.074 |
| inka1a | 0.169 | cox6b1 | -0.074 |
| elavl3 | 0.168 | aldh7a1 | -0.074 |
| dlb | 0.168 | acmsd | -0.074 |
| gpha2 | 0.167 | sult2st2 | -0.074 |
| kcnj11 | 0.164 | etnppl | -0.073 |
| rem2 | 0.164 | ftcd | -0.073 |
| neurod1 | 0.163 | cyp4v8 | -0.073 |
| si:dkey-10h3.2 | 0.159 | lgals2b | -0.073 |