ventral anterior homeobox 2
ZFIN 
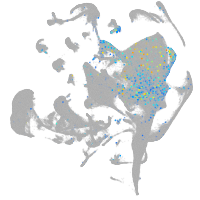
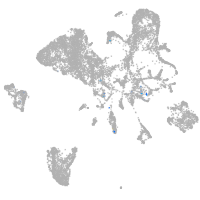
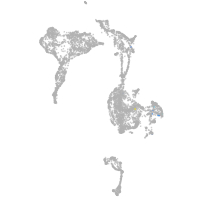
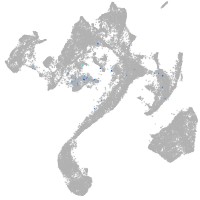
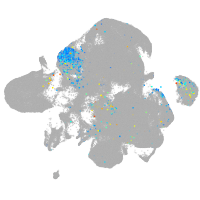
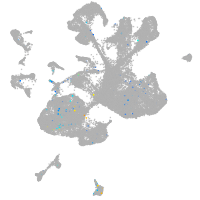
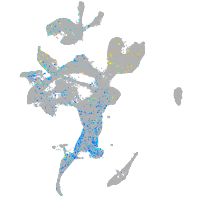
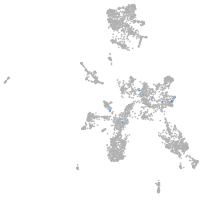
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| vax1 | 0.242 | bambia | -0.088 |
| ephb3 | 0.217 | efnb2a | -0.081 |
| foxi2 | 0.200 | foxg1b | -0.071 |
| aldh1a3 | 0.157 | ckbb | -0.067 |
| LO016987.2 | 0.156 | pvalb2 | -0.058 |
| rgmd | 0.155 | efnb1 | -0.057 |
| smoc2 | 0.152 | pvalb1 | -0.054 |
| ephb2b | 0.150 | nr2f2 | -0.049 |
| smoc1 | 0.130 | mdkb | -0.048 |
| six6a | 0.124 | calb2b | -0.047 |
| rdh10a | 0.124 | actc1b | -0.046 |
| cyp27c1 | 0.120 | si:ch211-152c2.3 | -0.045 |
| epha4b | 0.110 | hmgn6 | -0.044 |
| nr2f5 | 0.105 | gngt2b | -0.043 |
| zfp36l1a | 0.105 | foxg1d | -0.043 |
| bmpr1ba | 0.104 | syt5b | -0.041 |
| apcdd1l | 0.100 | myhz1.1 | -0.040 |
| foxd1 | 0.095 | si:dkey-16p21.8 | -0.039 |
| fzd5 | 0.095 | grk1b | -0.038 |
| her9 | 0.095 | tbx5b | -0.037 |
| her2 | 0.094 | pitx3 | -0.037 |
| ccnd1 | 0.090 | stm | -0.037 |
| mcm7 | 0.089 | apoa2 | -0.037 |
| vsx2 | 0.089 | hmgn3 | -0.037 |
| eno4 | 0.089 | mylz3 | -0.036 |
| sfrp1a | 0.089 | hbae3 | -0.036 |
| eomesa | 0.087 | hmx4 | -0.036 |
| c1qtnf12 | 0.086 | apoc1 | -0.036 |
| her12 | 0.085 | gnat2 | -0.036 |
| pax2a | 0.085 | cnrip1b | -0.036 |
| rrm1 | 0.084 | mylpfa | -0.036 |
| CABZ01005379.1 | 0.082 | hbbe1.3 | -0.035 |
| pcna | 0.082 | npdc1a | -0.035 |
| rx2 | 0.082 | si:ch73-256g18.2 | -0.035 |
| cyp1b1 | 0.081 | ak1 | -0.034 |