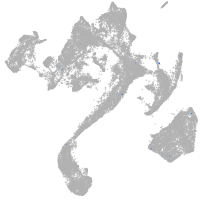
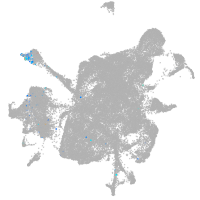
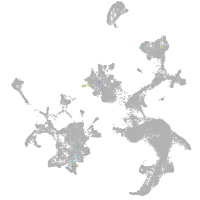
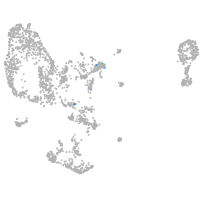
vesicle amine transport 1-like
ZFIN 
Other cell groups


Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| slc45a2 | 0.290 | ptmab | -0.110 |
| tyrp1b | 0.282 | si:ch73-1a9.3 | -0.102 |
| dct | 0.281 | ptmaa | -0.078 |
| pmelb | 0.279 | si:ch211-137a8.4 | -0.077 |
| pmela | 0.278 | gpm6aa | -0.074 |
| rab38 | 0.275 | hmgn6 | -0.071 |
| gpr143 | 0.274 | tuba1c | -0.069 |
| qdpra | 0.266 | cadm3 | -0.060 |
| tspan36 | 0.261 | marcksl1b | -0.060 |
| tspan10 | 0.260 | anp32e | -0.057 |
| tmem98 | 0.257 | hmgb1a | -0.056 |
| tyrp1a | 0.254 | rorb | -0.053 |
| col4a5 | 0.251 | tuba1a | -0.053 |
| oca2 | 0.251 | foxg1b | -0.051 |
| slc24a5 | 0.249 | h2afva | -0.050 |
| fabp11b | 0.249 | hmgn2 | -0.049 |
| agtrap | 0.249 | hmgb1b | -0.048 |
| mitfa | 0.246 | fabp7a | -0.048 |
| cracr2ab | 0.243 | nova2 | -0.046 |
| pah | 0.242 | snap25b | -0.046 |
| pnp4a | 0.239 | epb41a | -0.045 |
| sytl2b | 0.238 | marcksb | -0.045 |
| sparc | 0.237 | neurod4 | -0.045 |
| col4a6 | 0.236 | hmgb3a | -0.044 |
| slc7a9 | 0.235 | gapdhs | -0.044 |
| zgc:110591 | 0.231 | crx | -0.044 |
| bace2 | 0.231 | ndrg4 | -0.043 |
| gstp1 | 0.230 | si:ch1073-429i10.3.1 | -0.043 |
| FP085398.1 | 0.227 | baz2ba | -0.043 |
| mlphb | 0.227 | mex3b | -0.043 |
| tyr | 0.223 | hnrnpaba | -0.042 |
| rgrb | 0.222 | tmpob | -0.042 |
| LOC103910009 | 0.222 | sypb | -0.041 |
| abcg2d | 0.219 | neurod1 | -0.041 |
| rab32a | 0.219 | hmga1a | -0.041 |